-
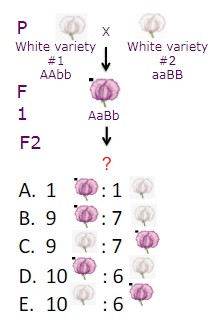
If you mate two AaBb F1 plants. What ratio of flower color do you predict to see in the F2?
B. 9:7
-
You study eye formation using Mexican cave-dwelling blind fish. You know that blindness is a trait controlled by multiple genes and inherited in a recessive manner.
A blind fish from a true-breeding line in one cave was crossed to a blind fish from a true-breeding line in another cave. If the mutation that causes blindness is in two different genes in the two fish, you should see:
A. None of the offspring are blind
B. 25% of the offspring are blind
C. 50% of the offspring are blind
D. 75% of the offspring are blind
E. All of the offspring are blind
A. None of the offspring are blind
-
If you mate two AaBb F1 plants. What ratio of flower color do you predict to see in the F2?
B
-
You study eye formation using Mexican cave-dwelling blind fish. You know that blindness is a trait controlled by multiple genes and inherited in a recessive manner.
A blind fish from a true-breeding line in one cave was crossed to a blind fish from a true-breeding line in another cave. If the mutation that causes blindness is in two different genes in the two fish, you should see:
A. None of the offspring are blind
B. 25% of the offspring are blind
C. 50% of the offspring are blind
D. 75% of the offspring are blind
E. All of the offspring are blind
A. None of the offspring are blind
-

You isolate 3 fish strains from different cave ponds, all the fish are blind because of autosomal recessive mutations. You mate the fish together (don’t worry about sex) and get the following results:
Fish strains #1 and #2 have mutations:
A. In the same gene
B. In different genes
A. In the same gene
-

You isolate two more blind fish strains (#4 and #5), cross them to #1, #2, and #3, and get the following results:
Based on these results, at least how many genes are working to produce sight?
3
-
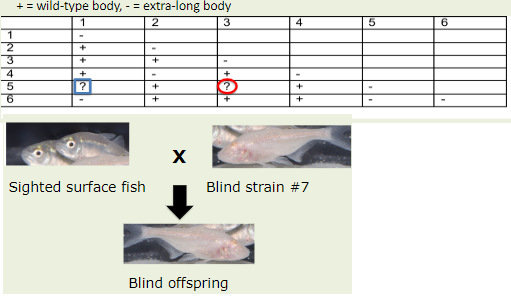
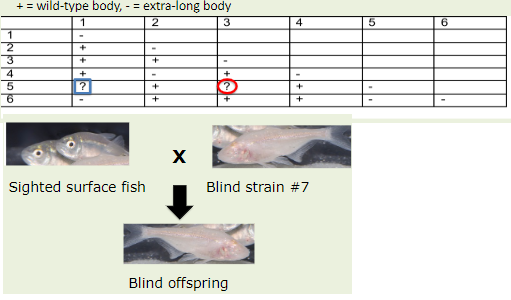
A researcher mutagenized a group of butterflies and isolated 6 butterflies with extra- long bodies. Each butterfly has a recessive mutation in only one gene. The researcher performed a complementation test using the 6 butterflies and got the following results.
Complete the chart.
Can we use the Strain #7 fish for complementation testing?
5-1: -
5-3: +
No
-

You cross strains 2 and 5, then have their offspring reproduce. Out of 80 offspring, how many are mutant and how many are wildtype?
45 wildtype
-
Randomly pick a base in the coding sequence and change it to any other base:
This change is copied during replication. Did this mutation create a new allele?
a) Yes
b) Yes, if the change is in the coding region of a gene
c) Yes, if the change is in the coding region of a gene and alters the amino acid sequence of a protein
d) No
a) Yes
-
The mutation that causes sickle cell disease is a type of:
A. Silent mutation
B. Missense mutation
C. Nonsense mutation
D. Frame shift mutation
B. Missense mutation
-
You invented a new shampoo called Lice-Gone that contains a chemical that kills lice. However, a recent study found that 53% of lice are resistant to the chemical in your shampoo. Which explanation is likely correct?
A) After being exposed to the shampoo, the lice needed to evolve resistance so they did not die a sudsy death.
B) Resistant strains of lice were always there—and are just more frequent now because all the non-resistant lice died a sudsy death.
A
-
You isolate a fly mutant line that never rests/sleeps and determine that this fly line has a dominant mutation in the gene party all night (pan).
In a wild-type fly, you only detect pan mRNA and protein in the adult brain. In the mutant line, pan mRNA and protein are found in the brain during all stages of fruit fly development.
This mutation is likely classified as a:
A. Gain of function mutation
B. Loss of function mutation
A. Gain of function mutation
-
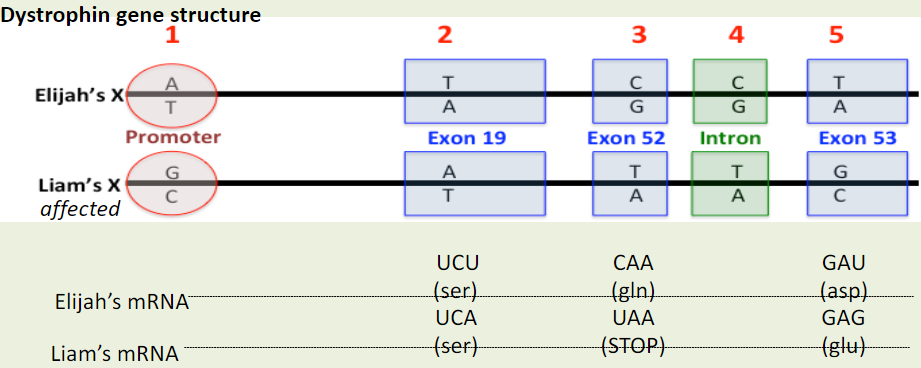
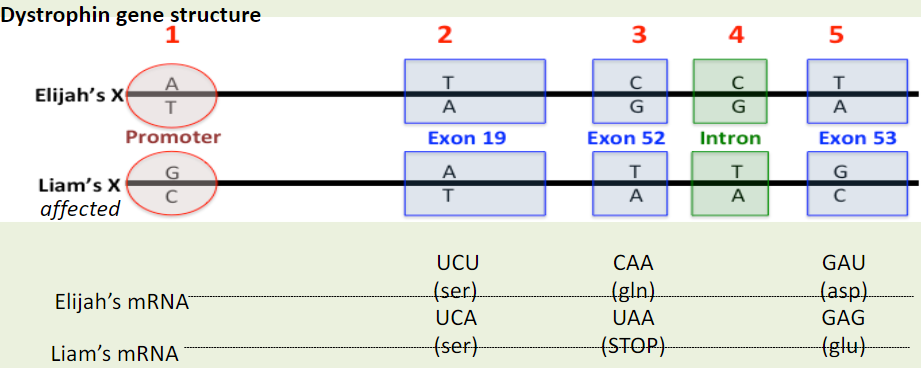
Is difference #5 a possible cause of DMD in Liam?
A. Yes
B. No
C. Maybe
Yes
-
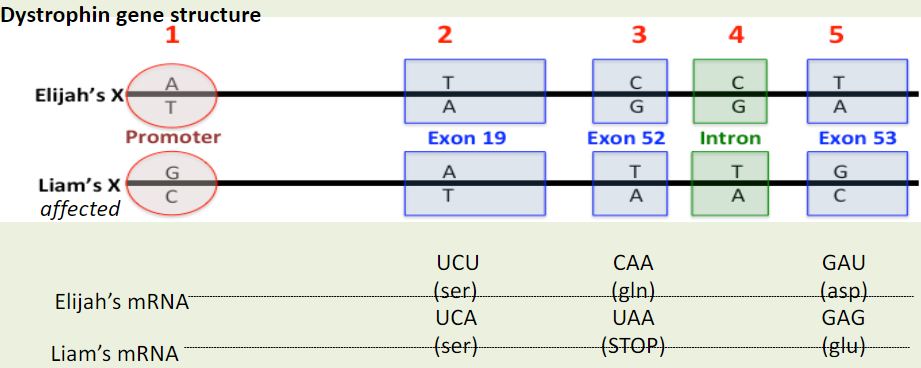
If difference #1 caused DMD, we would predict the mRNA levels in Liam to be __________ the mRNA levels in Elijah?
A. the same as
B. higher than
C. lower than
D. different in some way from
You determined that Elijah and Liam have the same mRNA levels. Would you conclude that difference #1 a likely cause of DMD in Liam?
D. different in some way from
No
-
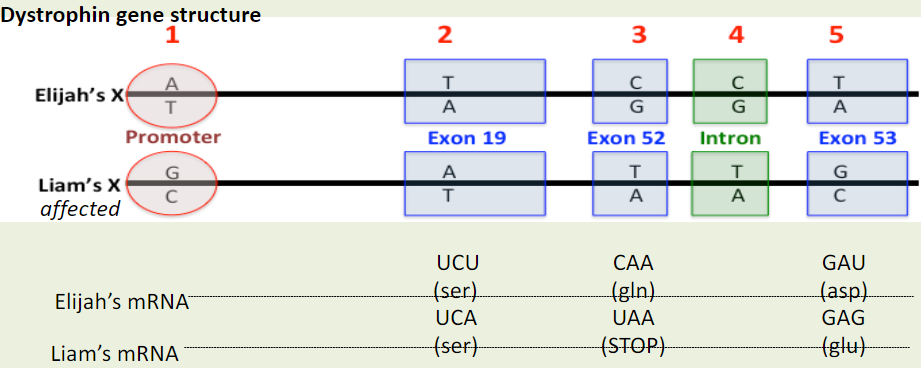
When DNA polymerase reaches the nucleotides encoding for the premature stop codon it will...
A. stop when it reaches the first nucleotide encoding for the premature stop codon.
B. stop when it reaches the last nucleotide encoding for the premature stop codon.
C. not be affected by this base change and will continue to read through the mutation.
What do you predict will be the effect of the premature stopcodon on mRNA size? It will result in:
A. a shorter mRNA in Liam.
B. a longer mRNA in Liam.
C. the same size mRNA in both Liam and Elijah.
C. not be affected by this base change and will continue to read through the mutation.
C. the same size mRNA in both Liam and Elijah.
-
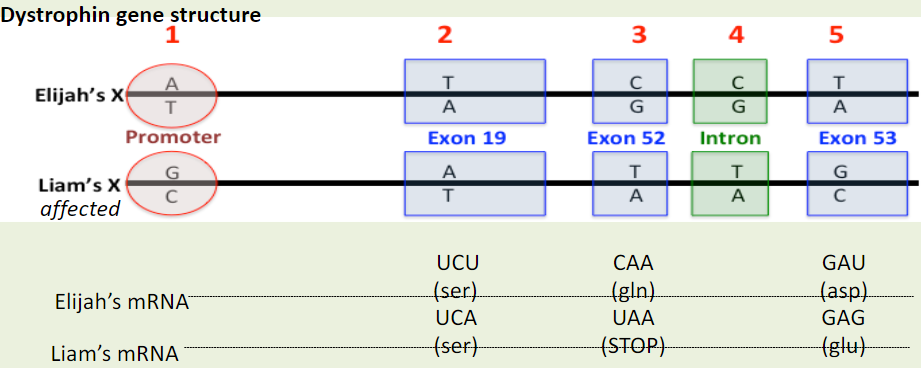
The above DNA sequence is being transcribed by an RNA polymerase (red square). The premature stop codon mutation in Liam’s DNA is indicated in blue.
What will the RNA polymerase do when it reaches the nucleotides encoding for the premature stop codon? It will:
A. stop when it reaches the first nucleotide encoding for the premature stop codon.
B. stop when it reaches the last nucleotide encoding for the premature stop codon.
C. not be affected by this base change and will continue to read through the change in the DNA.
What do you predict will be the effect of the premature stopcodon on protein size ? It will:
A. result in a smaller dystrophin protein in Liam.
B. result in a larger dystrophin protein in Liam.
C. result in the same size protein in both Liam and Elijah
C. not be affected by this base change and will continue to read through the change in the DNA.
A. result in a smaller dystrophin protein in Liam.
-
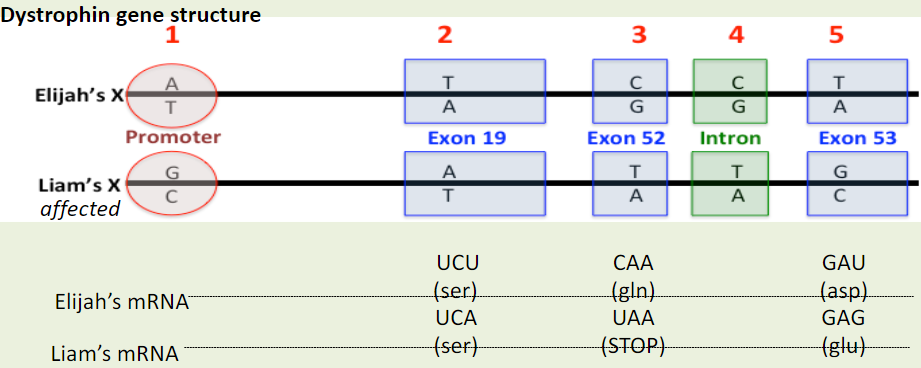
Which of the five nucleotide differences are mutations?
A. All five are mutations.
B. The three in the exons (difference #2, #3, #5)
C. The two in exons that change the amino acid sequence (differences #3, #5)
D. The one in the exon that causes DMD (difference #5)
A. All five are mutations.
-
Alleles: B = black; b = brown
A = ability to express black/brown pigment; a = inability to express black/brown pigment
What ratio of black to yellow dogs would you expect to get if you mated these two black dogs together:
AaBb and AaBB
A. All the dogs will be black
B. 4 black: 1 yellow
C. 3 black: 1 yellow
D. 2 black:1 yellow
E. 9 black: 4 yellow: 3 brown
C. 3 black: 1 yellow
-
A_ cats are solid white (the dominant white phenotype masks the phenotypes associated with the B/b and XE/Xe genes). aa cats can be other colors (orange, black, brown etc.)
How would you describe the type epistasis for the A gene over the B and X E genes for coat color in cats?
A. Recessive Epistasis
B. Dominant Epistasis
C. Double Recessive Epistasis
B. Dominant Epistasis
-
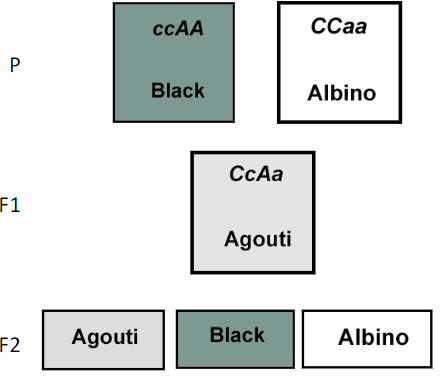
You cross two F1 agouti rats, what phenotypic ratio will you see in the F2?
C_A_ 9 Agouti
ccA_ 3 Black
C_aa 3 Albino
ccaa 1 Albino
-
What fragments would you get if you cut the same DNA with both EcoRI AND BamHI?
A. 600, 400, 850, and 150
B. 600, 250, and 150
C. 600 and 150
D. 850 and 400
B. 600, 250, and 150
-
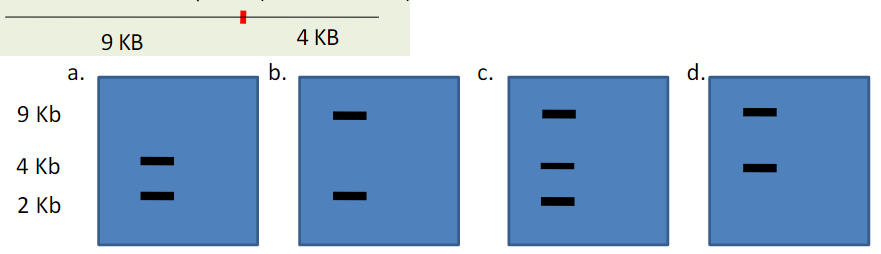
Cystic fibrosis is an autosomal recessive disease. You isolate the CFTR gene from an unaffected person and cut the DNA with the restriction enzyme NotI. NotI cuts the DNA into two pieces: a 9 KB and a 4 KB piece.
A common missense mutation in the CFTR gene introduces a new NotI restriction enzyme cut site. As a result the NotI enzyme cuts the CFTR gene into three pieces: 9 KB, 2 KB and 2 KB.
If you amplify DNA from a heterozygote, and perform Not1 digestion which of the following results would you expect?
C
-
What do the two bands in child’s lane represent?
A) two different alleles of the TPOX microsatellite on the same sister chromatid
B) two different alleles of the TPOX microsatellite on homologous chromosomes
C) two different alleles of the TPOX microsatellite on non-homologous chromosomes
B) two different alleles of the TPOX microsatellite on homologous chromosomes
-
Here is a pedigree of a family where some members have ataxia and a gel with the results of PCR amplification of the DI5S120 microsatellite. DI5S120 is tightly linked to the gene that causes ataxia when mutated.
Considering both the pedigree and the microsatellite data, what mode of inheritance is most likely?
A. Autosomal dominant
B. Autosomal recessive
C. X-linked dominant
D. X-linked recessive
These data are consistent with linkage between the gene that causes ataxia when mutated and D15S120. They imply that:
A. The D allele of D15S120 causes the disease phenotype
B. The D allele of D15S120 is linked to the gene that causes ataxia when mutated
C. Both A and B are correct
A. Autosomal dominant
B. The D allele of D15S120 is linked to the gene that causes ataxia when mutated
-
Here is a pedigree of a family where some members have Danon Disease and a gel with the results of PCR amplification of the CY3A microsatellite. CY3A is linked to the gene that causes Danon Disease when mutated.
What is the mode of inheritance?
A) autosomal dominant
B) autosomal recessive
C) X-linked dominant
D) X-linked recessive
Which CY3A allele is linked to the disease phenotype?
If II-2 and II-3 have another baby, what phenotype and gel banding pattern would you predict if the baby is a:
Boy:
Girl:
C) X-linked dominant
C
Boy: 0% chance of Danon Disease, either allele B or D of CY3A
Girl: 100% chance of Danon Disease, allele C of CY3A, and either allele B or D of
-
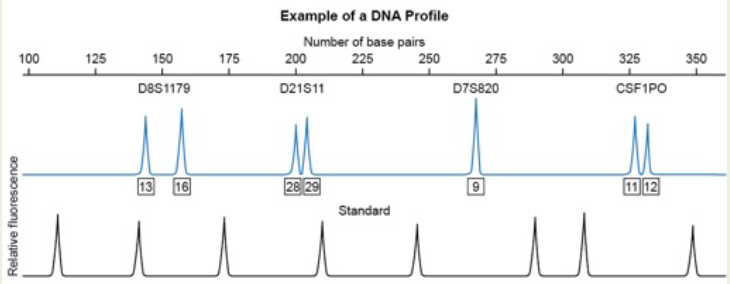
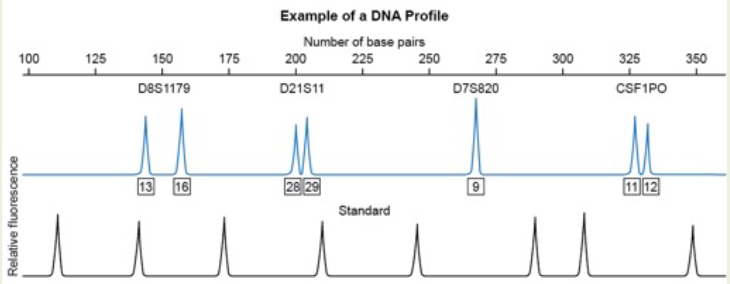
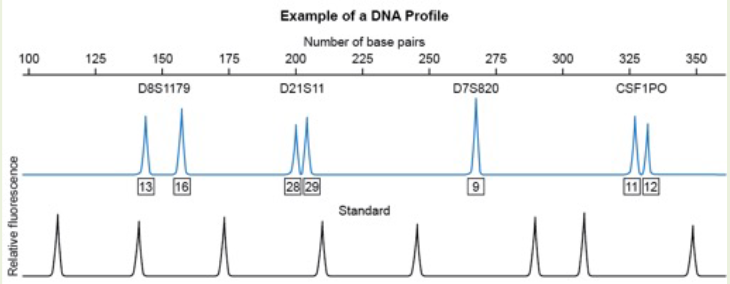
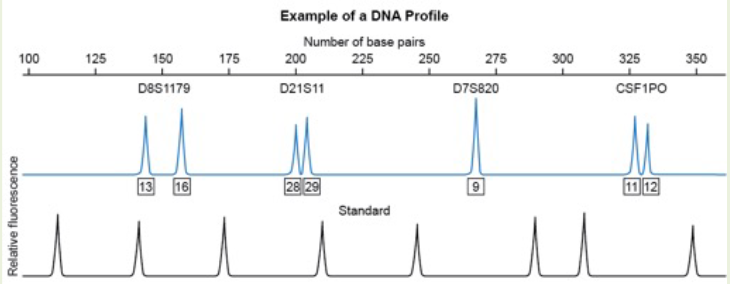
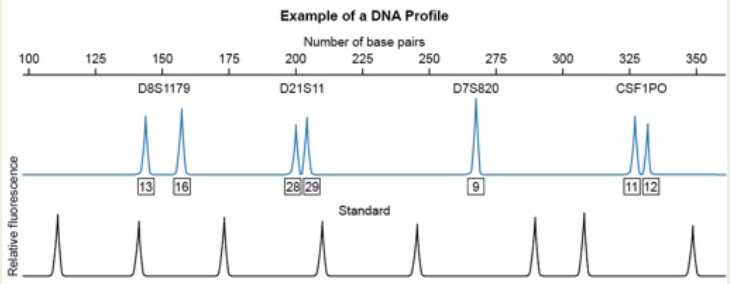
Based on this electropherogram, which microsatellite marker appears to be homozygous for this individual?
A. D8S1179
B. D21S11
C. D7S820
D. CSF1PO
C. D7S820
-
About 60% of the base pairs in a human DNA molecule are AT. If the human genome has 3.2 billion base pairs of DNA, about how many times will the following restriction sites be present?
a. BamHI (recognition sequence is 5’—GGATCC—3’)
b. EcoRI (recognition sequence is 5’—GAATTC—3’)
c. HaeIII (recognition sequence is 5’—GGCC—3’)
a. (0.2 x 0.2 x 0.3 x 0.3 x 0.2 x 0.2) x 3.2 x 10 9 = 460,800b. (0.2 x 0.3 x 0.3 x 0.3 x 0.3 x 0.2) x 3.2 x 10 9 = 1,036,800
c. (0.2 x 0.2 x 0.2 x 0.2) x 3.2 x 10 9 = 5,120,000
-
Which statements most accurately describe the first DNA strand produced by Taq polymerase during the PCR amplification process?
A. The Taq polymerase will keep extending past where the opposing primer pair is located
B. The Taq polymerase will stop where the opposing primer pair is located
A. The Taq polymerase will keep extending past where the opposing primer pair is located
-

In a mutant line of Arabidopsis, you identified a mutation in your gene of interest that results in the creation of an EcoRI restriction site that is not present in the wild type. You designed a Cleaved Amplified Polymorphic Sequence (CAPS) assay above.
In a homozygous mutant background, what band sizes would you expect after performing this assay and running the resulting digested PCR on a gel?
A. 600 bp bands only
B. 200 and 400 bp bands only
C. 200, 400 and 600 bp bands
B. 200 and 400 bp bands only
-
Your lab has decided to study albino fish. You know that albinism is controlled by multiple genes and is inherited in a recessive manner. You gather 10 mutant albino fish from various cave ponds in Mexico. You mate the fish together (don’t worry about sex) and get the following results:
If you cross strain 1 and strain 5, you would predict that the progeny will be:
a) 50% albino
b) 100% albino
c) 75% albino
d) 0% albino
e) 25% albino
Mutant 7 x Mutant 3 will result in:
a) you need to make more crosses to decide
b) +
c) -
b) 100% albino
a) you need to make more crosses to decide
-
You have established 3 homozygous strains of mice that are deaf.
Mouse strain 1: Deaf, phenotype caused by a recessive allele in a single gene.
Mouse strain 2: Deaf, phenotype caused by a recessive allele in a single gene.
Mouse strain 3: Deaf, phenotype caused by a recessive allele in a single gene.
True-breeding strain #1 mouse x true-breeding strain #2 mouse = hearing f1 progeny.
True-breeding strain #1 mouse x true-breeding strain #3 mouse = deaf f1 progeny.
True-breeding strain #2 mouse x true-breeding strain #3 mouse = ?
a) None of the mice will be deaf
b) 100% of the mice will be deaf
c) There is not enough information in this question to determine the
outcome
d) 50% of the mouse will be deaf
a) None of the mice will be deaf
-
You cross a strain #1 mouse to a strain #2 mouse and get F1 progeny. If you cross the F1 progeny together, what fraction of mice do you predict will be deaf in the F2 generation?
a) 7/16
b) 9/16
c) 3/16
d) 1/16
e) 4/16
a) 7/16
-
A researcher mutagenized a group of butterflies and isolated 6 butterflies with extra-long bodies. Each butterfly has a recessive mutation in only one gene. The researcher performed a complementation test using the 6 butterflies and got the following results.
+ = wild type body, - = extra-long body
What would be the result if a 5 butterfly were crossed with a 3 butterfly?
a) half of the offspring would have wild-type bodies and half of the offspring would have extra-long bodies
b) all the offspring would be wild-type
c) all of the offspring would have extra-long bodies
d) you need more information to answer this question
b) all the offspring would be wild-type
-
Mutations in the CFTR gene lead to the homozygous recessive disease CF.
Couple #1: baby is homozygous for a missense mutation, causing a serine to be made instead of a threonine. They are similar, but the change reduces the function of the CFTR protein.
Couple #2: baby is heterozygous for a nonsense mutation in the CFTR gene. The other allele is wild-type.
Couple #3: baby with missense mutation identified in couple #1’s baby and the nonsense mutation seen identified in couple 2’s baby.
Phenotype severity from least to most severe:
a) [LS] couple #2’s baby à couple #1’s baby à couple #3’s baby [MS]
b) [LS] couple #1’s baby à couple #2’s baby à couple #3’s baby [MS]
c) [LS] couple #3’s baby à couple #2’s baby à couple #1’s baby [MS]
d) [LS] couple #2’s baby à couple #3’s baby à couple #1’s baby [MS]
a) [least severe] couple #2’s baby à couple #1’s baby à couple #3’s baby [most severe]
-
A premature STOP codon can be created by:
a) a base substitution mutation
b) an insertion mutation
c) a deletion mutation
d) all of the above
d) all of the above
-
Mutations in the adenomatous polyposis of the colon (APC) gene predisposes a person to colorectal cancer. Below is the DNA nucleotide sequence of the APC gene on the coding strand from an unaffected individual and an individual who was diagnosed with colorectal cancer.
What type of mutation occurred in the individual that has colorectal cancer?
Note this sequence is from the middle of the APC gene. Please use the first 3 nucleotides for the first codon of this part of the APC gene.
a) missense
b) nonsense
c) frameshift
d) silent
b) nonsense
-
Here is the sequence around the translation initiation codon (AUG) of a human mRNA:
5’----AAGGGAUGGAACCUAGC....----3’
You have isolated a mutant version of the gene encoding this mRNA that contains a frameshift mutation. A one base pair insertion (base pair is underlined) occurs after the start codon (AUG). This change generates the mRNA shown below:
5’----AAGGGAUGGAAACCUAGC....----3’
This mutation will cause a premature stop in:
a) replication and transcription
b) transcription and translation
c) replication only
d) translation only
e) transcription only
d) translation only
-
One form of the autosomal recessive disease sickle cell anemia is caused by a mutation in the middle of the DNA sequence of the hemoglobin gene. If you look at the protein produced from this mutated sequence, and the protein is the wild-type length, what type of mutation is most likely?
a) silent
b) frameshift
c) nonsense
d) missense
d) missense
-
A single base deletion mutation of “A” nucleotide has occurred at the start codon of the APC2 gene and is passed on to the next generation. This mutation has been named APC2del. In the offspring that has inherited APC2del, this allele will most likely be:
a) not translated
b) not transcribed
c) not replicated
d) none of the above
d) none of the above
-
RNA interference (RNAi) is a method where you introduce double-stranded RNA in a cell or organism to silence (turn off) a specific gene expression. You use RNAi to target the Mma2 gene in the worm C. elegans. You predict that you will get a:
a) wild-type phenotype
b) a loss of function phenotype
c) a gain of function phenotype
b) a loss of function phenotype
-
For any microsatellite DNA marker, what is the maximum number of bands you can see for a diploid individual on a gel?
2
-
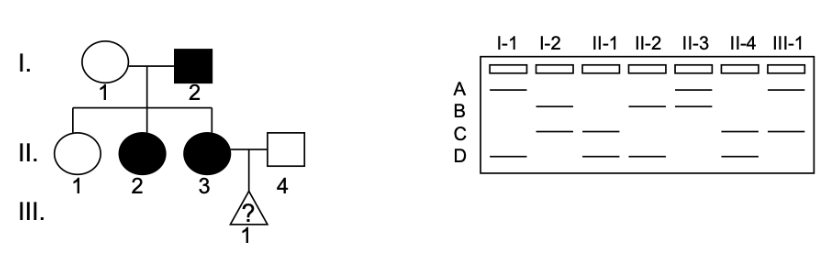
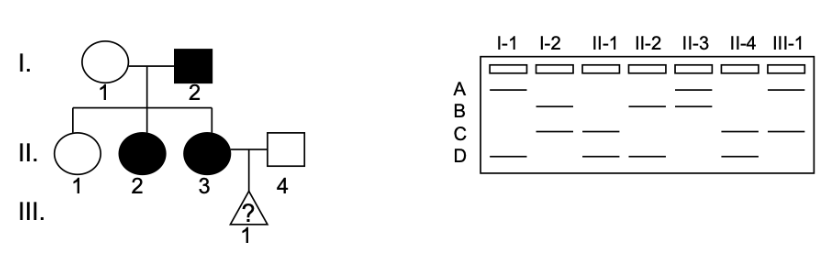
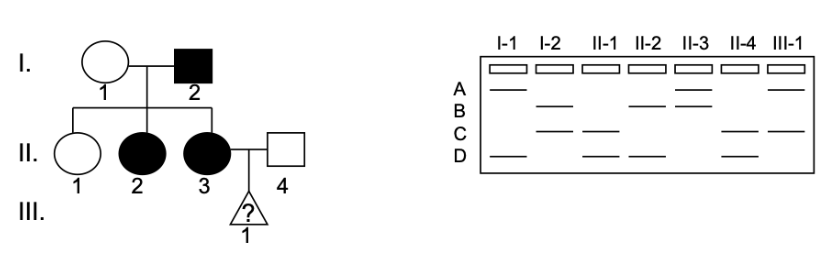
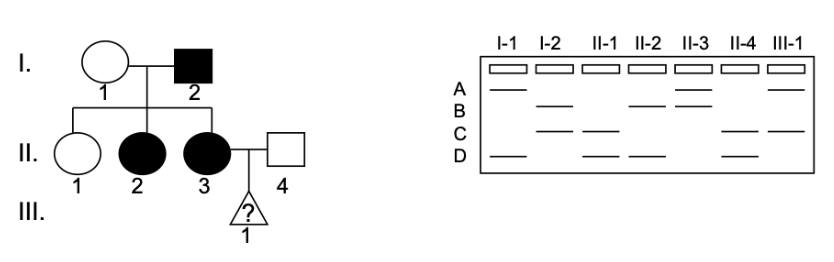
A family has late onset Machado-Joseph Disease. Shown is the microsatellite closely linked to the disease trait. No recombination between the gene of interest and the microsatellite has occurred in this family. III-1 is a fetus. The sex and the disease status of this fetus are unknown.
How is Machado-Joseph Disease inherited?
a) autosomal recessive
b) autosomal dominant
c) X-linked recessive
d) X-linked dominant
Which of the following is most likely true about III-1:
a) III-1 is a boy who does not have the disease
b) III-1 is a boy who has the disease
c) III-1 is a girl who does not have the disease
d) The sex of III-1 cannot be determined but they have the disease
e) The sex of III-1 cannot be determined but they do not have the disease
f) III-1 is a girl who has the disease
b) autosomal dominant
e) The sex of III-1 cannot be determined but they do not have the disease
-
A woman who does not have sickle cell anemia has a sister with sickle cell anemia. Both of her parents do not have sickle cell anemia.
This non-affected woman has children with a man from sub-Saharan Africa who does not have sickle cell anemia. However, there is no information about whether this disease runs in his family.
Use the Hardy-Weinberg equation with the information that approximately 96% of the people do not have the autosomal recessive disease sickle cell anemia to determine the probability that the man is a carrier.
What is the probability that they will have a child with sickle cell anemia?
a) 0.04
b) 0.25
c) 0.05
d) 0.15
c) 0.05
-
A population of water snakes is found on an island in Lake Erie. Some of the snakes are banded and some are unbanded; the banding phenotype is autosomal recessive. The frequency of banded snakes on the island is 0.4.
There are 500 total snakes on the island. How many snakes are heterozygous for the banding allele? Round to the nearest whole snake!
a) 300
b) 200
c) 233
d) 184
e) 316
c) 233
-
Tongue rolling
R = ability to roll tongue
r = cannot roll tongue
In a population of 100RR, 100Rr and 100rr
What is the frequency of allele R?
a) 0.33
b) 0.50
c) 0.66
d) 0.75
e) 0.25
b) 0.50
-
In sub-Saharan Africa, approximately 96% of the population do not have the autosomal recessive disease sickle cell anemia. What is the frequency of the autosomal dominant HbA allele?
a) 0.98
b) 0.8
c) 0.96
d) 0.2
e) 0.04
b) 0.8
-
The MN blood type is conferred by co-dominant alleles LM and LN.
In the Maldives, 1% of the population has the blood group N. What is the expected frequency of individuals that have the heterozygous MN blood group, assuming Hardy Weinberg equilibrium?
a) 0.45
b) 0.02
c) 0.01
d) 0.90
e) 0.18
e) 0.18
-
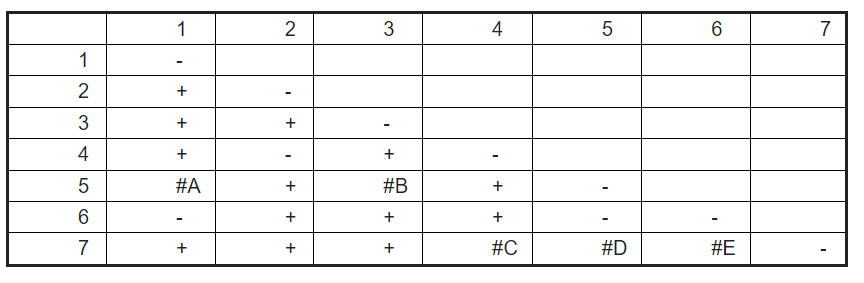
A researcher mutagenized a group of butterflies and isolated 7 butterflies with extra-long bodies. Each butterfly has a recessive mutation in only one gene. The researcher performed a complementation test using the 7 butterflies and got the following results.
+ = wild-type body,
- = extra-long body
Based on the data available, would expect complementation (+) or non-complementation (-) in the blank labeled A, B, C, D, E?
-, +, +, +, +
-

What is the frequency of the A allele? (assume these frogs are diploid)
80%
-
In a frog population, the frequency of allele A = 0.8, and the frequency of allele a = 0.2 .
What percentage of frogs in the next generation will be AA?
What percentage will be aa?
What percentage will be Aa?
AA = 64%
aa = 4%
Aa = 32%
For Aa, remember that two possibilities exist: Aa and aA
Remember 2pq
-

Tay-Sachs (TS) is inherited in an autosomal recessive manner bythe “a” allele.
1 in 2500 births are infants with TS.
What is the frequency of the “a” allele?
0.02
-

Tay-Sachs (TS) is inherited in an autosomal recessive manner bythe “a” allele.
1 in 2500 births are infants with TS.
What is the probability that an unaffected Jewish woman with nohistory of TS is a carrier (Aa) if you assume the population is atH-W equilibrium?
If the same unaffected US Jewish woman has a child with a manwho is a carrier (Aa), what is the chance that their child willhave TS (aa)?
0.04;
1%
-
In cats, all-white color is dominant over not all-white. In a populationof 1000 cats, 190 are all-white cats.
Assuming that cats are diploid and the population is in Hardy-Weinberg equilibrium, what is the frequency of the all-white allele inthis population?
0.1

