Enzymes I and II (The Living Cell)
Learning objectives: Classify the functions of enzymes and why they are needed Describe the features of an enzyme active site Explain the key factors that contribute to rate enhancement and specificity in enzyme reactions. Measurement of Vmax and Km from enzyme kinetics. Describe what is meant by competitive and non-competitive enzyme inhibition and how they may be distinguished List the properties of allosteric enzymes Recognise that many anti-tumour and antibacterial agents act by inhibiting specific enzymes
-
Outline of Proteins and their functions:
• Digestion: carbohydrates, fats, proteins
• Blood clotting: fibrin clot catalysed by thrombin
• Defence-immune system-activation of complement
• Movement: muscle actomyosin is an ATPase
• Nerve conduction: membrane pumps for Na+, K+, Ca++
-
Inherited diseases such as phenylketonuria, glycogen storage disease and Tay-Sachs disease can be detrimental; What do these diseases do? (1 point each)
• phenylketonuria-cannot convert Phe to Tyr.
• glycogen storage disease-cannot mobilise glucose
• Tay-Sachs disease-defect in processing a membrane ganglioside, which results in neuronal damage and death
-
Enzymes are drug targets; what do antibiotics such as penicillin do?
Inhibit cell wall synthesis
-
Enzymes are drug targets; What do anti-inflammatory agents such as aspirin do?
Blocks prostaglandin synthesis
(a group of lipids with hormone-like actions that your body makes primarily at sites of tissue damage or infection that influences blood clotting.)
-
Enzymes are drug targets; What do anticancer drugs such as methotrexate do?
It is a folate analogue which interferes with synthesis of DNA precursors
-
Enzymes increase reaction rate by how much?
Increase reaction rate by up to10 billion-fold!
-
State 5 key points about enzymes:
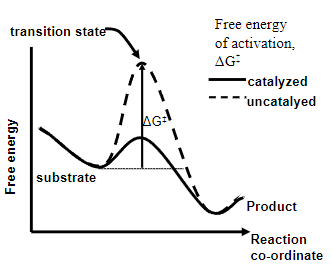
• Show specificity
• Unchanged at end of reaction
• Do not alter reaction equilibrium
• Enzymes speed up reaction by decreasing the free energy of activation of the reaction
•Have active sites that bind substrate(s)
-
What is Enzyme Catalysis?
The acceleration of a chemical reaction by a biological catalyst, the enzyme
-
Enzymes reduce ΔG‡ and speed up the reaction by using what?
Enzyme-substrate binding energy
-
What are 7 factors that are responsible for enzyme catalysis?
• To bring molecules together in active site
• To constrain substrate movement
• To strain particular bonds in the substrate
• To stabilise positive and negative charges in transition state
• To exclude water from the active site-make reaction go faster
• To provide a reaction pathway of lower energy
• Use cofactors
-
Picture outlining how lysozyme is a good 'straining' enzyme
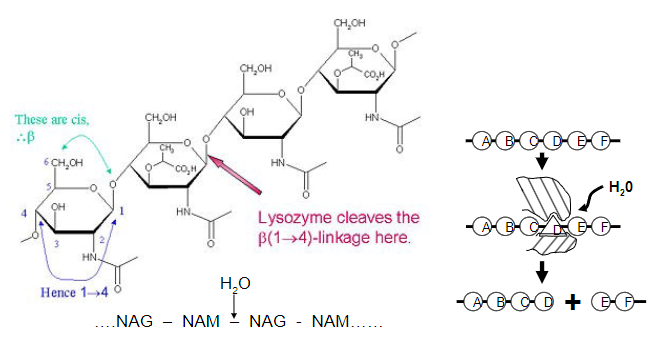
-Polysaccharide cleavage causes bacterial rupture and death
-
Enzymes have active sites; What are they?
• The active site is a 3-D cavity or cleft that binds substrate(s)with specificity through electrostatic, hydrophobic, hydrogen bonding and van der Waals interactions.
-
Where does the evidence for active sites come from?
• X-ray crystallography (usually done at synchrotrons-Diamond, Oxford)
• Kinetic studies of enzyme activity (simpler and quicker than crystallography)
-
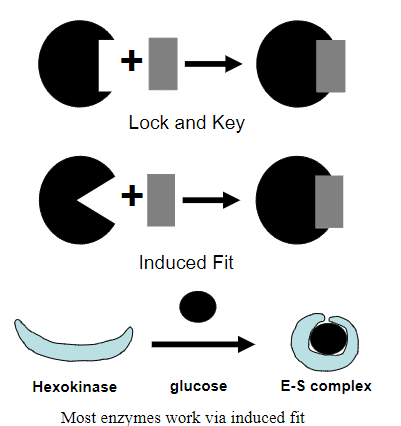
Picture outlining the 'Lock and Key' mechanism vs the 'induced fit' mechanism:
-
A piece of evidence for induced fit binding is...?
Substrate glucose to Enzyme Hexokinase
-
A piece of evidence for active sites is…?
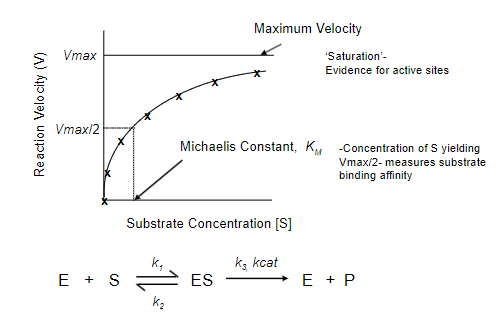
Michaelis-Menten Kinetics
-
What does the Michaelis-Menten Model describe?
Describes the relationship between the rate of an enzyme-catalysed reaction and the concentration of substrate.
-
What is the Michaelis-Menten equation? What does each part represent?
![The Michaelis-Menten equation is expressed as (picture):where:V0 is the initial reaction velocity.Vmax is the maximum reaction velocity.[S] is the substrate concentration.Km is the Michaelis constant, representing the substrate concentration at which the reaction velocity is half of Vmax.](/flashcards/cardimage2/3f7bdd7/046/8046574_back.png)
The Michaelis-Menten equation is expressed as (picture):
where:
V0 is the initial reaction velocity.
Vmax is the maximum reaction velocity.
[S] is the substrate concentration.
Km is the Michaelis constant, representing the substrate concentration at which the reaction velocity is half of Vmax.
-
The Lineweaver-Burk equation is the inverse of the Michaelis-Menten equation; Can you state the Lineweaver-Burk equation and label it in the format y=mx+c
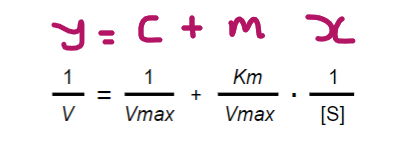
-
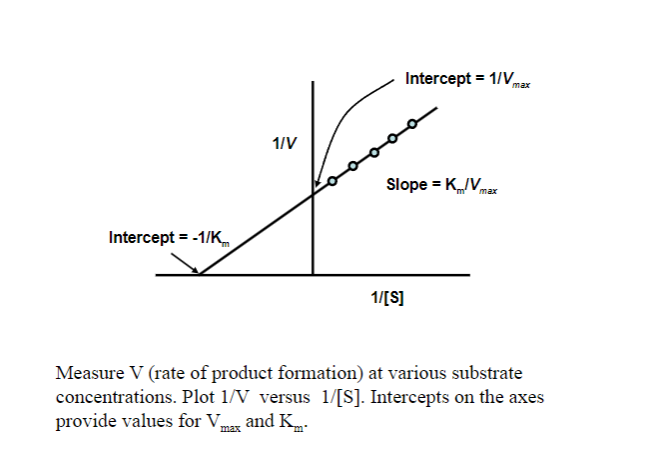
Picture outlining Obtaining Vmax and Km (use a double reciprocal) plot:
-
What is the k1 Constant?
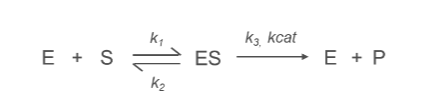
k1:
Represents the rate constant for the formation of the enzyme-substrate complex (ES) from the free enzyme (E) and substrate (S).
E+S→ES
-
What is the k2 Constant?
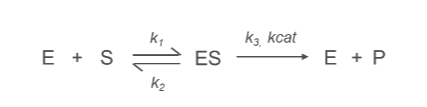
k2:
Denotes the rate constant for the conversion of the enzyme-substrate complex (ES) into product (P) and free enzyme (E).
ES→E+P
Describes the efficiency of the catalytic step, specifically the breakdown of the enzyme-substrate complex.
-
What is the k3/kcat Constant?
kcat (Turnover Number):
Represents the rate constant for the conversion of enzyme-substrate complex (ES) into product (P) and free enzyme (E).
ES→E+P
Reflects the maximum catalytic efficiency of the enzyme, indicating how many substrate molecules an enzyme can convert into product per active site per unit time
-
What is the turnover number?
Max no of substrate molecules handled per activesite per second
-
What is Km?
A measure of substrate binding affinity-in μM (micromolar) to mM range (millimolar)
-
Inhibitor effects on kcat and Km can distinguish what?
Competitive vs non-competitive inhibitors-valuable in drug design work
-
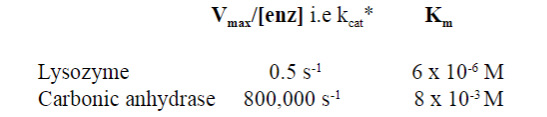
Look at this picture: What is the kcat for most enzymes?
kcat for most enzymes is ~10 s -1
-
Inhibitor I compete with substrate S for binding to the enzyme active site, forming what?
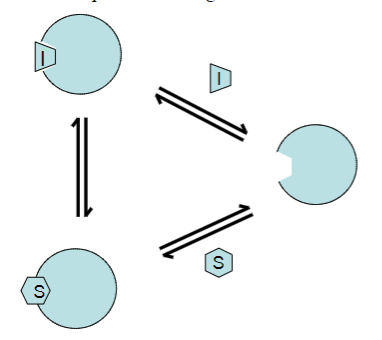
An Inactive EI complex
-
In the presence of a competitive inhibitor, does Km increase or decrease? Why?
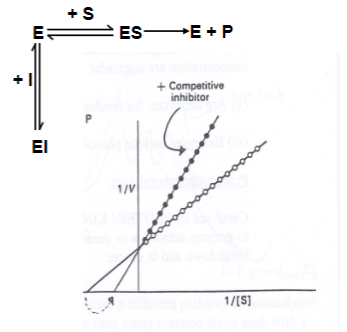
Km is increased (it takes more substrate to achieve Vmax/2).
However, Vmax is unaltered, as the effects of the inhibitor can be competed out at high substrate concentrations.
-
Enzyme activity is regulated in cells by 4 mechanisms; What are they?
• Control of gene expression-enzyme amount
• Compartmentation: sequences in enzyme polypeptidechain target enzyme to ER, mitochondrion, nucleus etc
• Allosteric regulation: a regulatory molecule (acting at apocket distinct from the active site) changes the enzymeconformation to influence the active site and decrease (or insome cases, increase) enzyme activity. Controls the fluxof material through a metabolic pathway.
• Covalent modification of enzyme. Change enzyme shape and activity-e.g. phosphorylation
-
Picture outlining how Feedback inhibition regulates metabolic pathways:
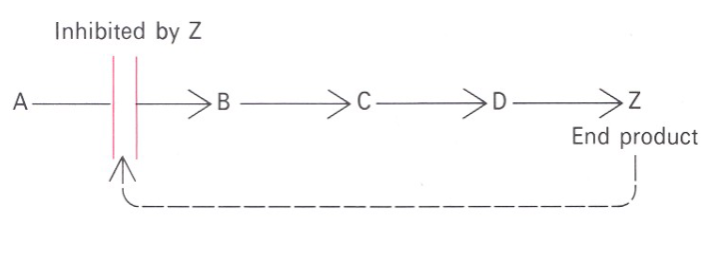
-
Picture outlining ATCase-R6C6, catalytic and regulatory sitesare on different subunits:
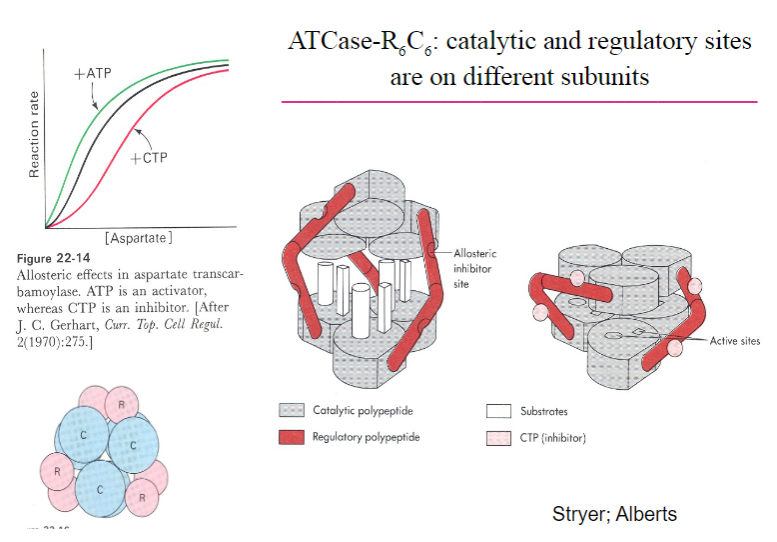
-
A summary of Allosteric enzymes:
• Multisubunit complexes• Regulatory sites and catalytic sites ondifferent subunits• Regulation occurs via conformational changes• Exhibit non-Michaelis-Menten kinetics:V vs S plots are sigmoidal• Involved in feedback inhibition of metabolic pathways
-
What enzyme coils DNA in bacteria? And what is the other enzyme that uncoils it?
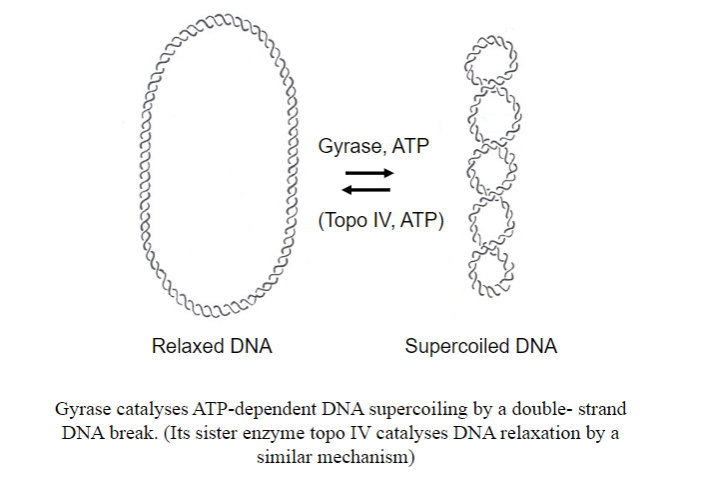
-
What antibiotic inhibits gyrase?
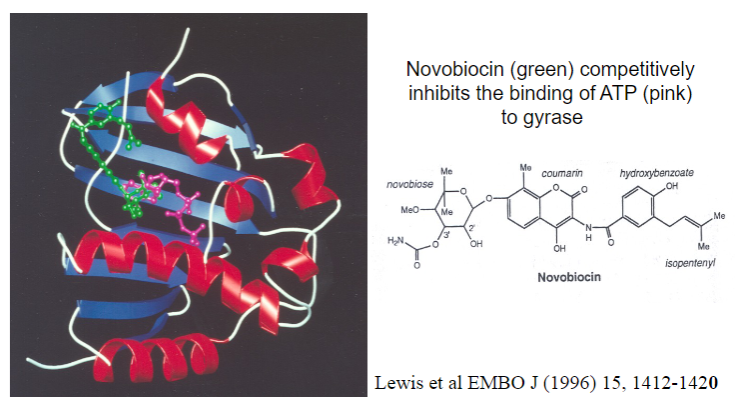
Novobiocin
-
What is a competitive inhibitor of the enzyme dihydrofolate reductase, that makes DNA bases?
Methotrexate-an anticancer folate analogue
-
In enzyme evolutions acts to maximise....?
Catalytic efficiency
-When evolution has sped up the 'chemical' step to the greatest degree
-
So called 'perfect' enzymes have reaction rates limited by what?

Diffusion
-
What is considered the most 'perfect' enzyme?
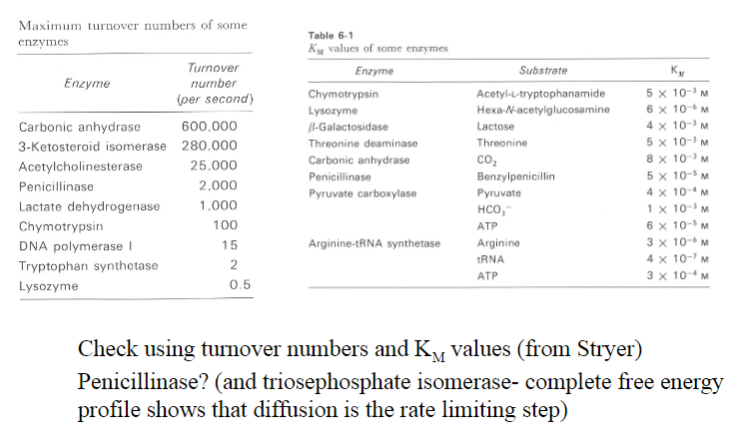
Carbonic anhydrase
-
What are peptide bonds resistant to?
Hydrolysis
-
What activates the catalytic serine?
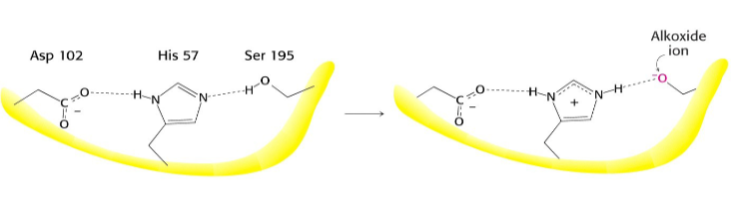
A charge-relay system by proton withdrawal
-
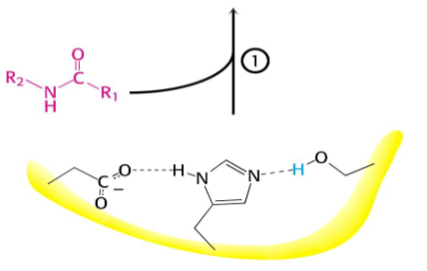
What occurs after this step?
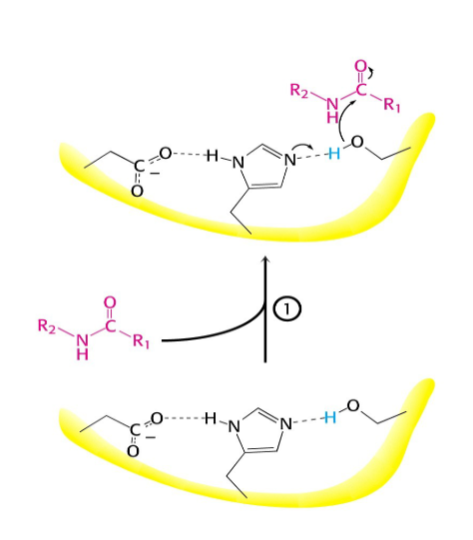
-
Serine proteases have a very reactive serine, what does this serine do?
Attacks the peptide bond to form an acyl-enzyme
-
Picture outlining how Peptide hydrolysis by serine proteases is a two-step process:
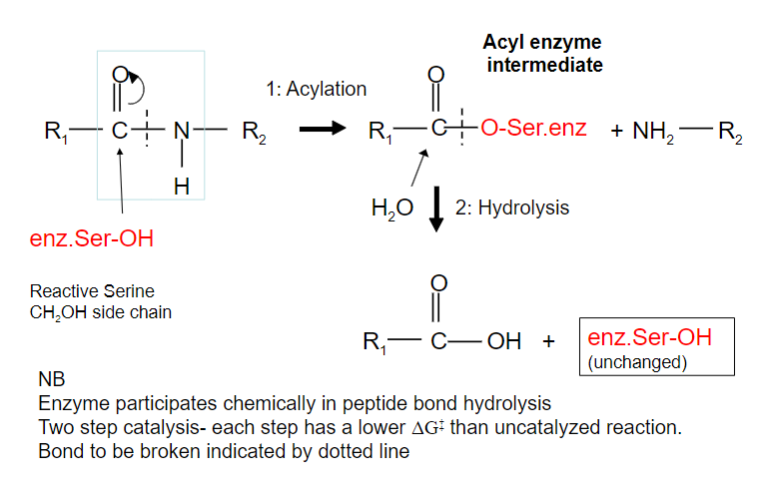
-
Picture demonstrating how Serine proteases hydrolyse protein peptide bonds with sequence specificity:
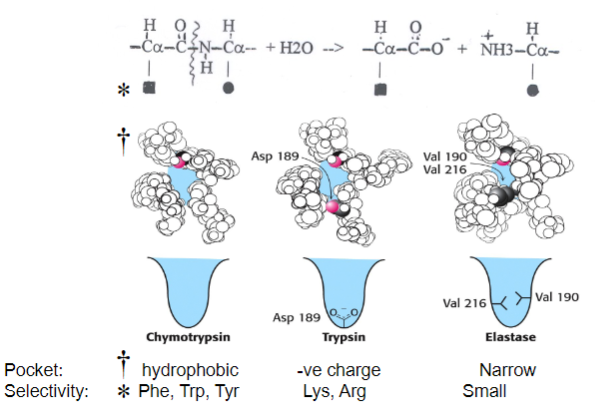
-
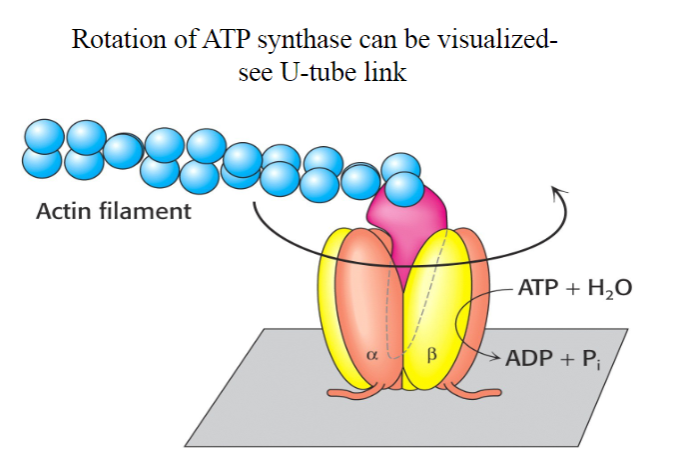
How many active sited does Rotary ATP Synthase have? What activates it?
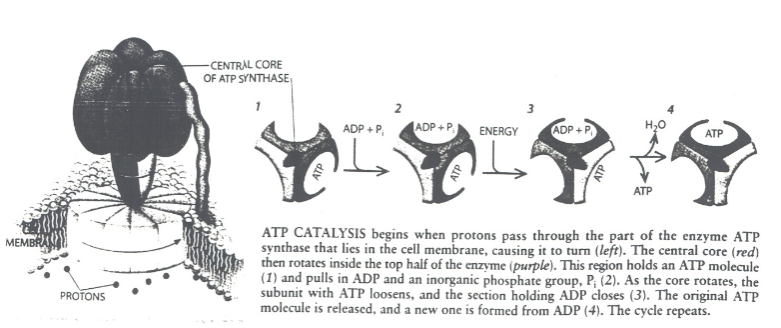
-3
-A rotating spindle
-
Do bacteria have their own rotary ATP synthase?
Yes
-
What does the antituberculosis drug bedaquiline do?
Kills Mycobacterium tuberculosis by inhibiting its ATP synthase.
-
What is the Topo II enzyme?
An enzyme clamp that unlinks daughter chromosomes
-
How does Topo II work? (1 line)
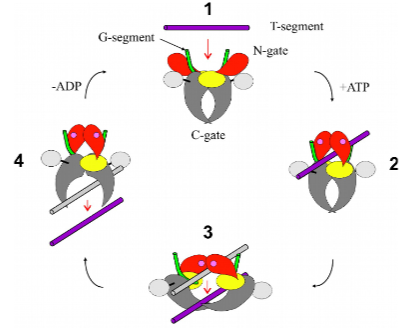
Cross, a DNA segment from one chromosome through a transient ds-DNA break in another (a temporary or short-lived interruption in both strands of the DNA double helix).
-
What are the roles of actin and myosin in muscle contraction?
Actin:
Forms thin filaments in muscle fibres.
Has binding sites for myosin heads.
Undergoes conformational changes during muscle contraction.
Myosin:
Forms thick filaments in muscle fibres.
Possesses myosin heads that interact with actin.
Utilizes ATP hydrolysis to generate force and movement.
Cross-bridges between actin and myosin contribute to muscle contraction.
-
How is muscle contraction regulated in skeletal muscle?
Neural Stimulation:
Initiated by nerve impulse, releasing acetylcholine at neuromuscular junction.
Acetylcholine binds to sarcolemma receptors on the muscle cell membrane.
Action Potential Propagation:
Action potential travels along sarcolemma and into transverse tubules (T-tubules).
Calcium Release:
Action potential triggers release of calcium ions from sarcoplasmic reticulum (SR).
Troponin and Tropomyosin Interaction:
Calcium binds to troponin, causing conformational changes.
Tropomyosin shifts, exposing myosin-binding sites on actin.
Cross-Bridge Formation:
Myosin heads bind to exposed actin sites, forming cross-bridges.
Contraction:
ATP hydrolysis and power strokes lead to sliding of actin and myosin filaments, inducing muscle contraction.
Relaxation:
Calcium actively transported back into SR
-
How is muscle contraction regulated in smooth muscle?
Calcium Signalling:
Calcium influx through cell membrane channels and intracellular stores.
Calmodulin Activation:
Calcium binds to calmodulin, forming the calcium-calmodulin complex.
MLCK Activation:
Calcium-calmodulin activates Myosin Light Chain Kinase (MLCK).
Myosin Phosphorylation:
MLCK phosphorylates myosin, enabling interaction with actin.
Cross-Bridge Formation:
Myosin-actin cross-bridge formation initiates contraction.
MLCP:
Myosin Light Chain Phosphatase (MLCP) dephosphorylates myosin, promoting relaxation.
Latch State:
Some smooth muscles maintain force with low energy consumption (latch state) due to sustained myosin phosphorylation.
-
Why are chymotrypsin, trypsin and elastase known as ‘serine proteases’? What are their biological functions?
Why Named Serine Proteases:
Contain serine residues in their active sites.
Biological Functions:
Chymotrypsin, trypsin, and elastase play roles in protein digestion.
Chymotrypsin cleaves at hydrophobic residues.
Trypsin cleaves at basic residues.
Elastase cleaves at small, uncharged residues
-
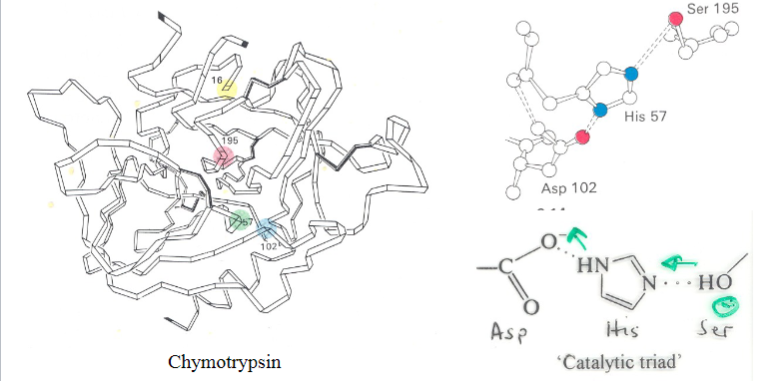
What do you understand by the term ‘catalytic triad’ and what is its function in serine proteases
Definition:
Consists of three amino acids (Serine, Histidine, Aspartate) in the active site.
Function:
Coordinates the catalytic reaction by stabilizing reaction intermediates.
Serine acts as a nucleophile in substrate cleavage.
-
How is the proton gradient set up across the inner mitochondrial membrane? How does it store energy for use by the ATP synthase?
Set-Up:
Generated during electron transport chain in oxidative phosphorylation.
Protons pumped from matrix to intermembrane space.
Energy Storage:
Potential energy stored in the form of a proton gradient.
Released during ATP synthase-catalysed proton movement back to matrix.
-
How do topologically interlocked chromosomes arise in cells and how are they unlinked by topo II?
Formation:
Can result from DNA replication or recombination errors.
Examples include catenanes or knot-like structures.
Unlinking by Topoisomerase II:
Topoisomerase II introduces transient double-strand breaks and resolves interlocked structures.
-
What is the definition of a ‘catalytically perfect’ enzyme? Does this property apply to all enzymes?
Definition:
Achieves catalysis at the diffusion-limited rate.
Applicability:
Ideal concept; practically unattainable due to limitations such as substrate diffusion.
-
Hydrolysis of peptide bonds in proteins is thermodynamically favourable. Why then are peptide bonds in proteins stable to hydrolysis in water?
Thermodynamic Favourability:
Hydrolysis releases energy; however, the rate is slow without enzymes.
Stability in Proteins:
Protein structure and the surrounding environment protect peptide bonds from spontaneous hydrolysis.
-
Describe four examples of molecular machines and their functions in cells.
Examples and Functions:
Ribosome:
Protein synthesis.
ATP Synthase:
ATP production during oxidative phosphorylation.
DNA Polymerase:
DNA replication.
Helicase:
Unwinds DNA strands during replication and repair.

