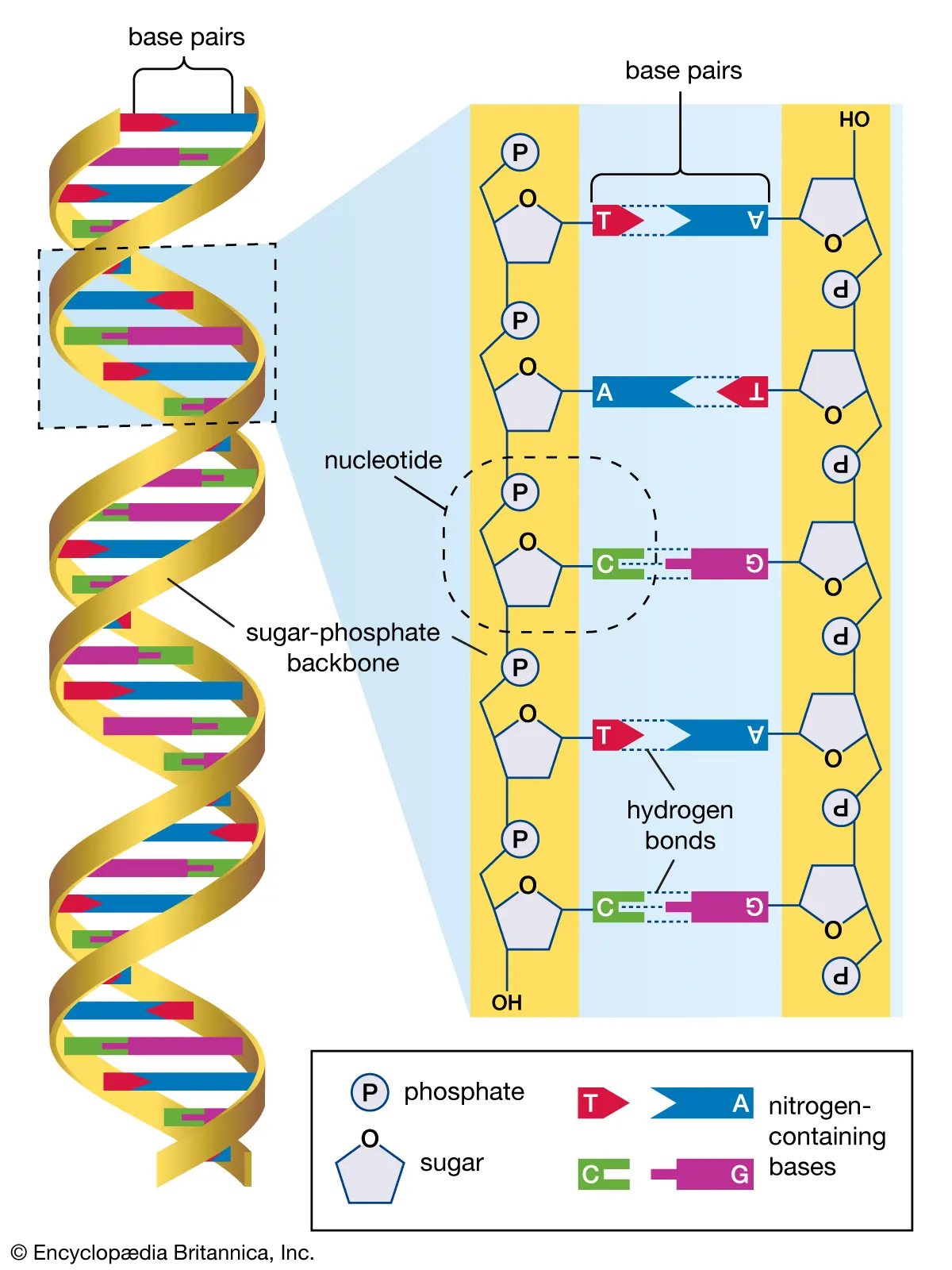
DNA Sequencing (Genomics)
SDL: DNA Sequencing During the last 30 years, a revolution in our understanding of the human genome has taken place. This is primarily because of our ability to perform modern molecular techniques, to reveal not only the sequence of the human genome, but also to understand the molecular function of genes through our ability to decipher the genetic code base by base. The human genome project was a multinational collaborative effort that took over 13 years requiring multi-billion dollar funding and generating the first “complete” human genome by 2006. It is a project that was facilitated largely by a single method, dideoxy chain termination sequencing. A method first described in 1977 by Fred Sanger and others. This technique although now superseded by next generation sequencing for whole genome sequencing, it remains the gold standard for confirming suspected mutations and many other focused applications in medicine and molecular biology. Learning Outcomes On successful completion of the lecture, students should be able to: Describe in detail the process of Dideoxy sequencing, also known as Sanger sequencing Similarity to PCR but with specific differences The detail of the four stages of the reaction itself Size separation of the reaction products by gel electrophoresis Base-calling by software Describe uses of Dideoxy sequencing in healthcare and research
-
What is dideoxy chain termination commonly referred to as? (1)
Sanger Sequencing
-
Who developed the dideoxy chain termination method and when? (2)
Developed by Fred Sanger in 1977.
Nobel Prize winner.
-
How has the dideoxy chain termination method evolved over time? (2)
The method remains the same as originally developed.
Technology improvements and the technique has become semi-automated
-
What are the advantages of dideoxy chain termination sequencing? (3)
Robust with a low error rate.
Highly reliable, still considered a gold standard technique.
Widely used due to its accuracy.
-
What is the role of the ABI 3730 in automated DNA sequencing? (2)

Automates dideoxy chain termination sequencing on a large scale.
Uses robotics for sample preparation.
-
What are the capabilities of the ABI 3730 sequencer? (3)

Read length of up to 900 base pairs.
99.95% accuracy.
Handles 48 or 96 samples simultaneously.
-
How many samples can the ABI 3730 process per day? (1)

>1000 samples per day.
-
What is the limitation of the ABI 3730 sequencing system? (2)

Only separates labelled DNA and determines the sequence.
Requires manual preparation of samples before sequencing.
-
What major project was the ABI 3730 used for? (1)
It was used to sequence the Human Genome.
-
How many bases of sequence did the ABI 3730 produce for the Human Genome project? (1)
23 thousand million bases (23 Gbases).
-
How long did it take and how much did it cost to complete the Human Genome sequencing using ABI 3730? (2)
It took 13 years and cost $2.7 billion.
-
What are the key steps in sequencing by dideoxy chain termination? (4)
Template preparation
Enzymatic sequencing reaction
Size separation of products by capillary electrophoresis
Detection of reaction products and readout of sequence
-
How does DNA polymerase contribute to dideoxy chain termination sequencing? (1)
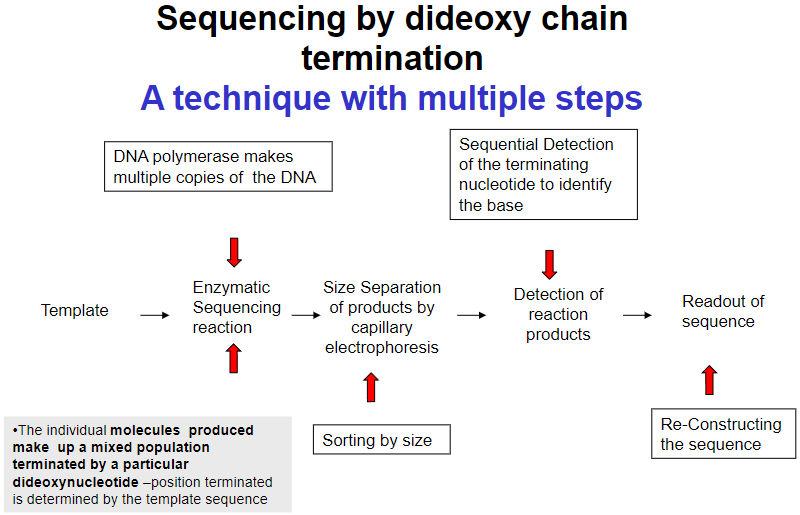
DNA polymerase makes multiple copies of the DNA during the sequencing reaction.
-
How are the products sorted in dideoxy chain termination sequencing? (1)
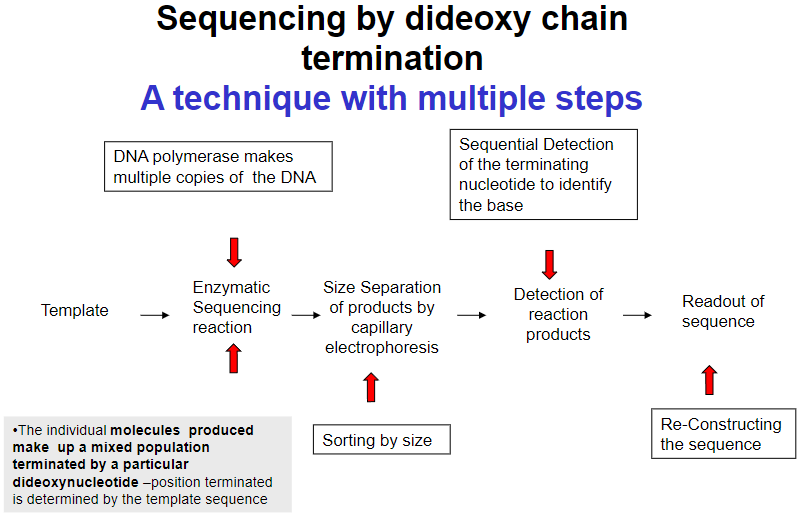
Products are sorted by size through capillary electrophoresis.
-
How is the terminating nucleotide detected in sequencing? (1)
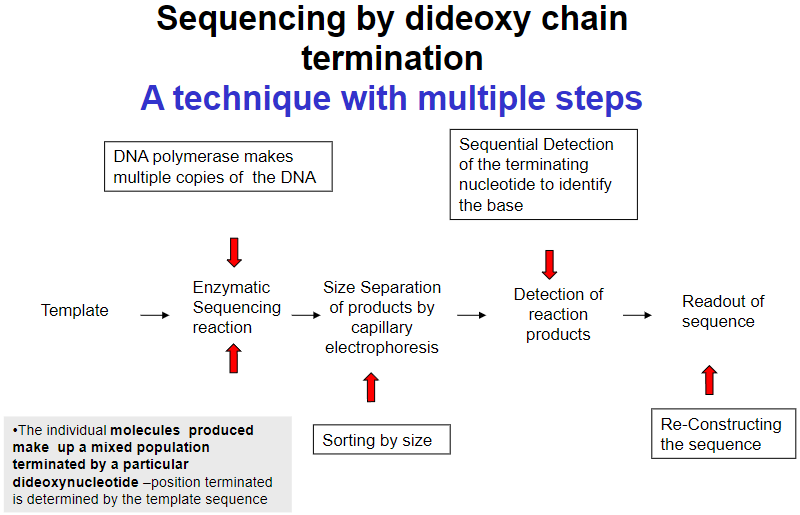
Sequential detection of the terminating nucleotide identifies the base.
-
How is the DNA sequence reconstructed in dideoxy chain termination sequencing? (1)
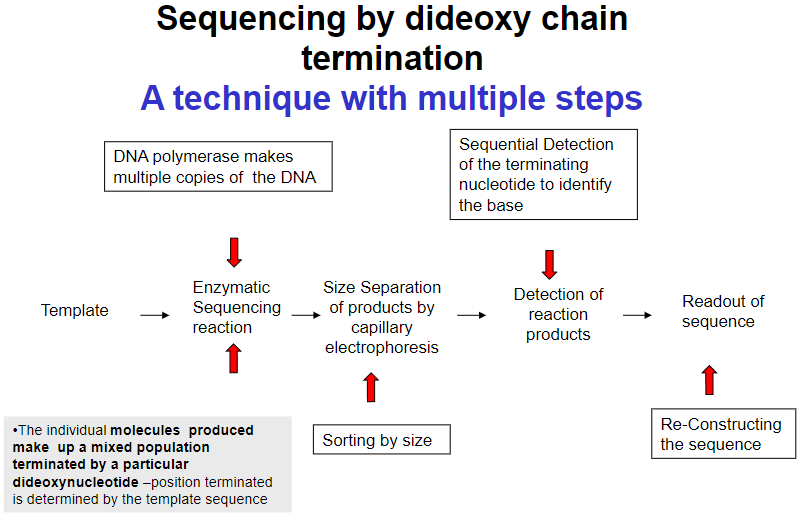
The individual molecules produced form a mixed population, and the terminated position is determined by the template sequence, which helps in reconstructing the sequence.
-
How is dideoxy chain termination sequencing similar to PCR? (1)
Both use DNA polymerase in the reaction process.
-
What is a key difference between dideoxy chain termination sequencing and PCR? (1)
Dideoxy sequencing uses only a single forward primer, while PCR typically uses both forward and reverse primers.
-
Is amplification exponential in dideoxy chain termination sequencing? (1)
No, amplification is limited, not exponential.
-
When cycling is performed in dideoxy chain termination sequencing, what type of polymerase is usually used? (1)
A thermostable polymerase is usually used for cycling.
-
Why is a thermostable polymerase necessary in some protocols of dideoxy sequencing? (1)
A thermostable polymerase is necessary because some protocols cycle through repeated temperatures.
-
What is the first step in the dideoxy chain termination sequencing reaction? (1)
Strand separation to create single-stranded DNA templates.
-
What happens after strand separation in the dideoxy sequencing reaction? (1)
Annealing of the primer to the single-stranded DNA template.
-
What occurs during the extension step of dideoxy chain termination sequencing? (1)
DNA polymerase extends the primer, adding nucleotides to the growing DNA strand.
-
What is the final step in the dideoxy chain termination sequencing reaction? (1)
Chain termination, where the incorporation of a dideoxynucleotide prevents further extension of the DNA strand.
-
What is the first step in sequencing using DNA polymerase? (1)
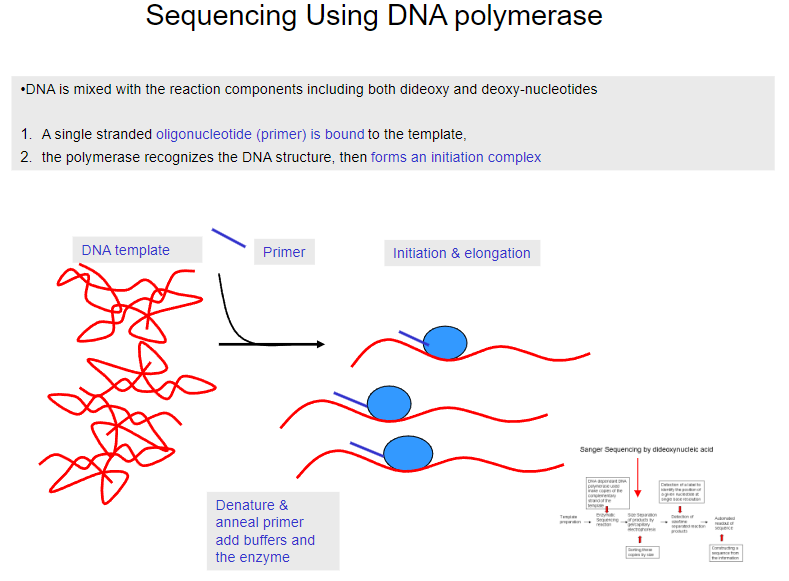
DNA is mixed with reaction components, including both dideoxy and deoxy-nucleotides.
-
What happens after the DNA is mixed with the reaction components in sequencing? (1)
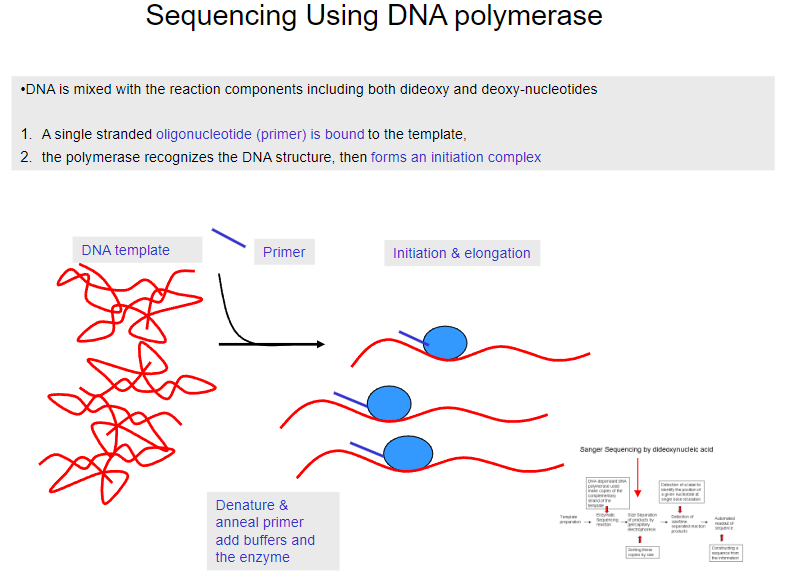
A single-stranded oligonucleotide (primer) binds to the template DNA.
-
What does the polymerase do in the sequencing process? (1)
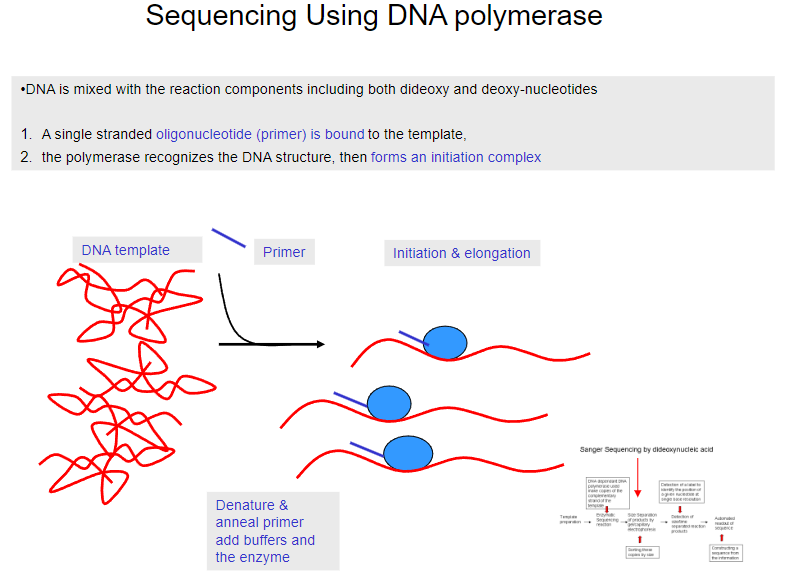
The polymerase recognizes the DNA structure and forms an initiation complex.
-
What happens after the primer binds to the template DNA? (1)
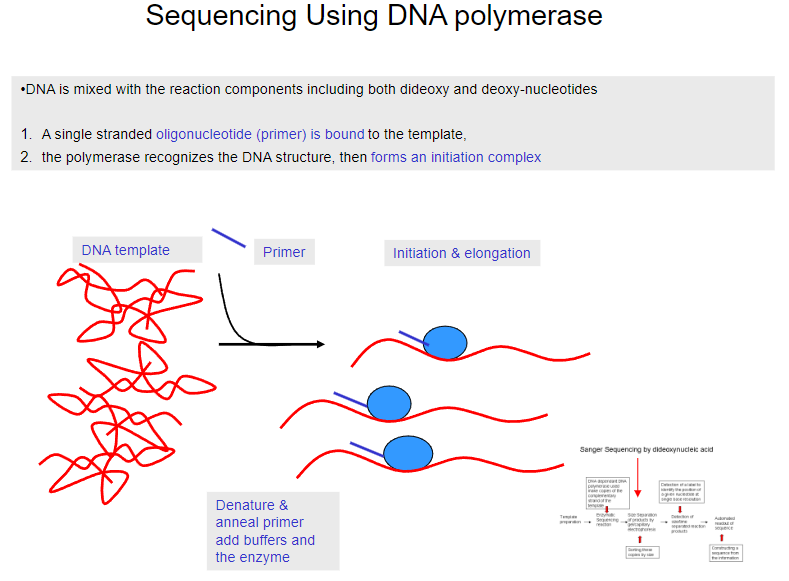
The primer is denatured and annealed, and buffers and the enzyme are added.
-
What is required for DNA-dependent DNA polymerase to function? (4)
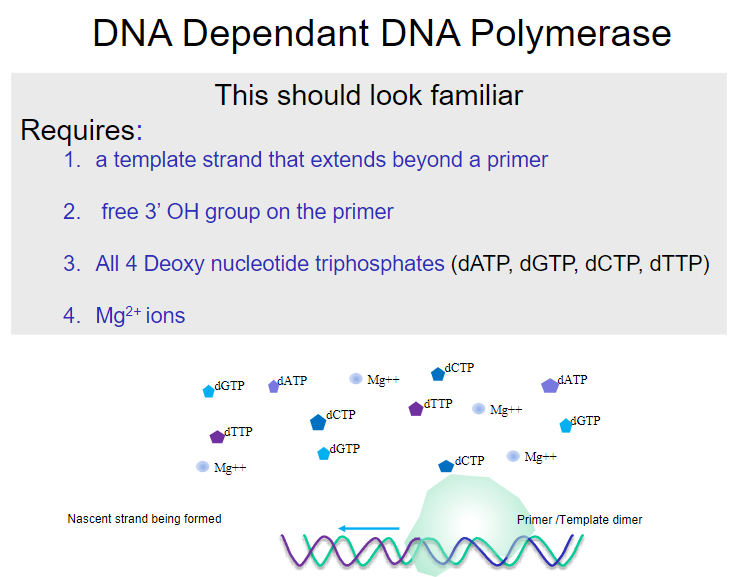
A template strand that extends beyond a primer.
A free 3’ OH group on the primer.
All four deoxy nucleotide triphosphates (dATP, dGTP, dCTP, dTTP).
Mg2+ ions.
-
What is the nascent strand in DNA polymerase activity? (1)
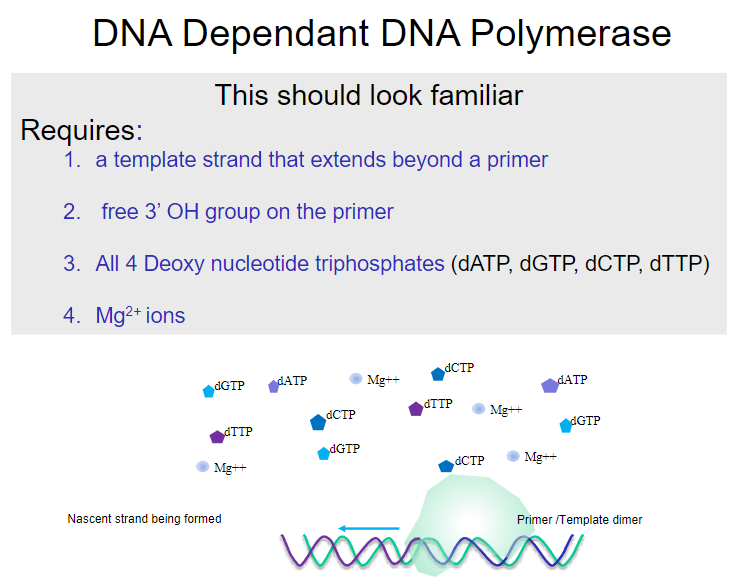
The nascent strand is the newly synthesized DNA strand formed during the polymerase reaction.
-
Picture demonstrating A closer look at the elongating strand:addition of dNTP (base):
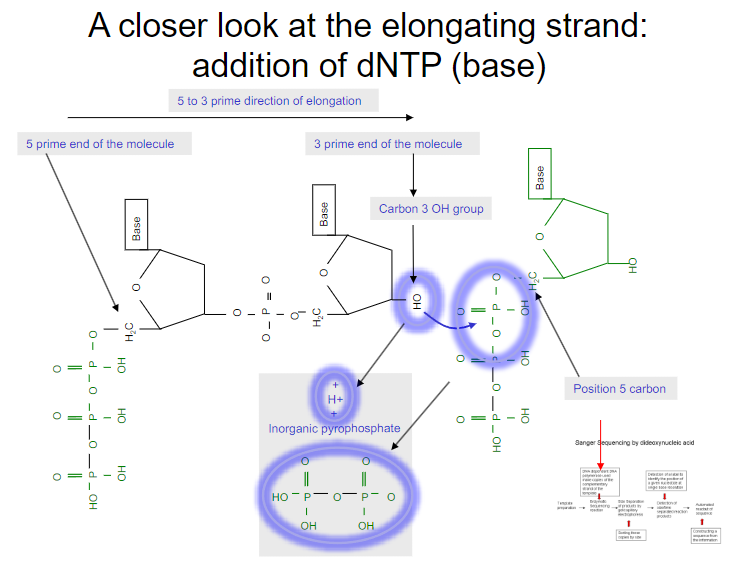
-
What are the requirements for chain termination in DNA sequencing? (5)

A template strand that extends a primer, forming a partial duplex.
A free 3’ OH group on the primer.
All four deoxy nucleotide triphosphates (dATP, dGTP, dCTP, dTTP).
All four dideoxy nucleotide triphosphates (ddATP, ddGTP, ddCTP, ddTTP).
Mg2+ ions.
-
What happens during chain termination in DNA sequencing? (1)
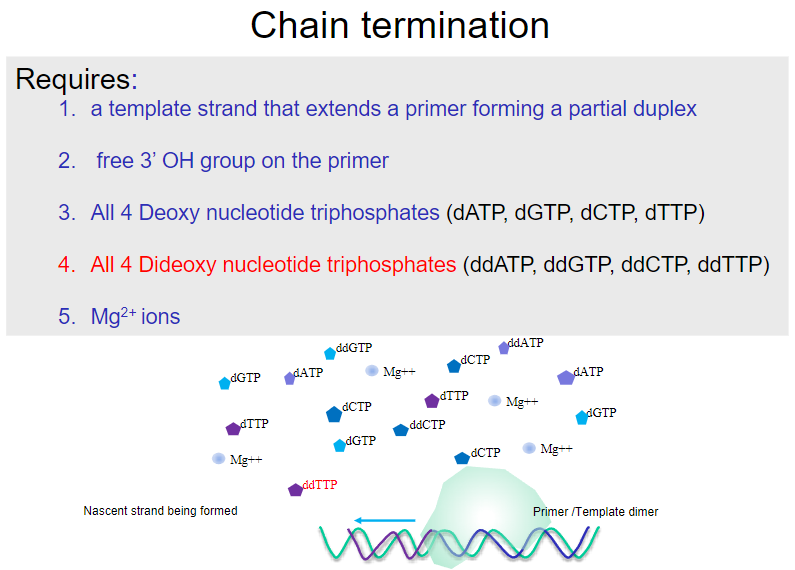
Chain termination occurs when a dideoxynucleotide (ddNTP) is incorporated into the growing strand, which lacks a 3’ OH group, preventing further elongation.
-
How is DNA elongation terminated in DNA sequencing? (1)
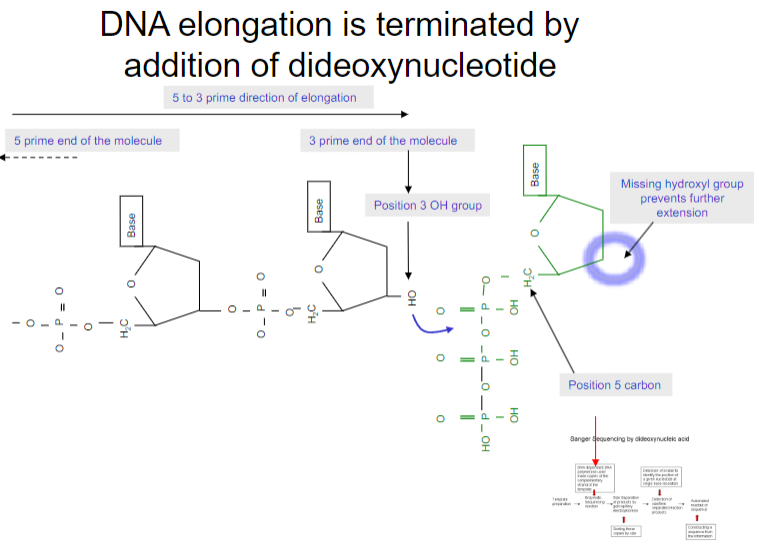
DNA elongation is terminated by the addition of a dideoxynucleotide (ddNTP), which lacks a 3’ OH group, preventing further elongation of the DNA strand.
-
How do dideoxynucleotides prevent DNA elongation? (1)

Dideoxynucleotides prevent elongation by lacking a 3’ OH group, which is essential for the addition of the next nucleotide, thereby terminating the DNA strand elongation.
-
What happens during sequencing using DNA polymerase? (2)
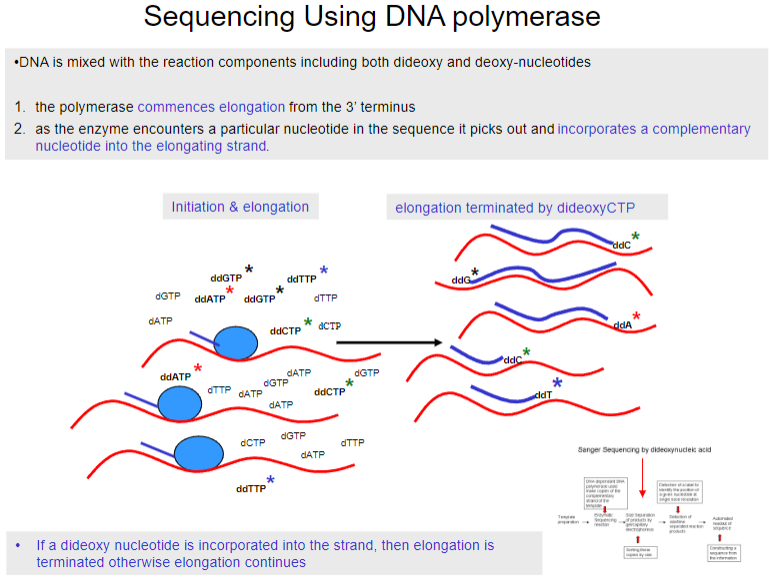
The polymerase begins elongation from the 3’ terminus.
As the enzyme encounters a particular nucleotide in the sequence, it selects and incorporates the complementary nucleotide into the elongating strand.
-
How do reaction products vary in length in sequencing by dideoxy chain termination? (2)

Products terminated by ddCTP represent all positions in the sequence where cytosine (C) occurs.
The population of molecules produced includes all possible positions in the sequence, from the same point to the end, for each of the four labelled dideoxy nucleotides.
-
How does the size of reaction products help determine the sequence in dideoxy chain termination? (2)

Reaction products vary in length due to termination by ddNTPs.
Ordering these molecules by size allows us to determine the sequence of the newly synthesized strand.
-
How is size separation of reaction products achieved in dideoxy chain termination sequencing? (4)
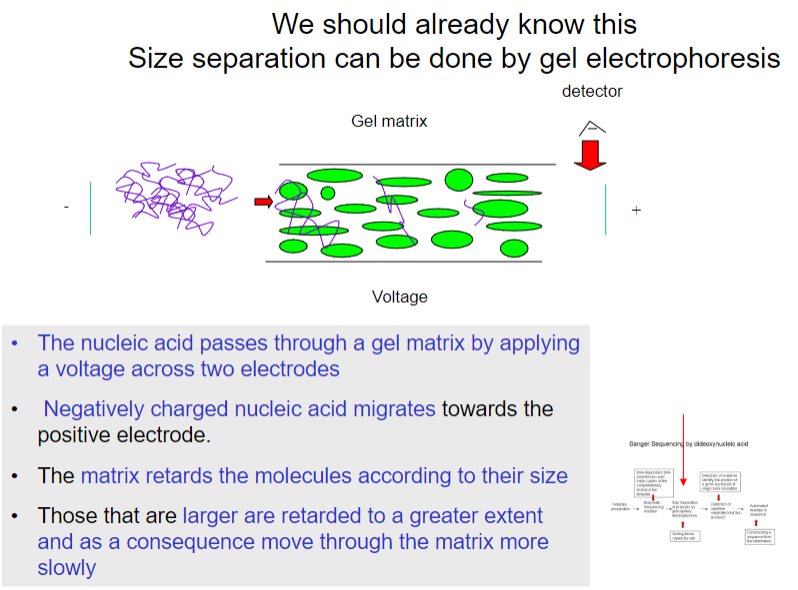
Size separation is done by gel electrophoresis, where nucleic acids pass through a gel matrix with a voltage applied across electrodes.
The negatively charged nucleic acids migrate toward the positive electrode.
The gel matrix retards molecules based on size.
Larger molecules move more slowly due to greater retardation.
-
How is the DNA sequence determined in dideoxy chain termination sequencing? (2)
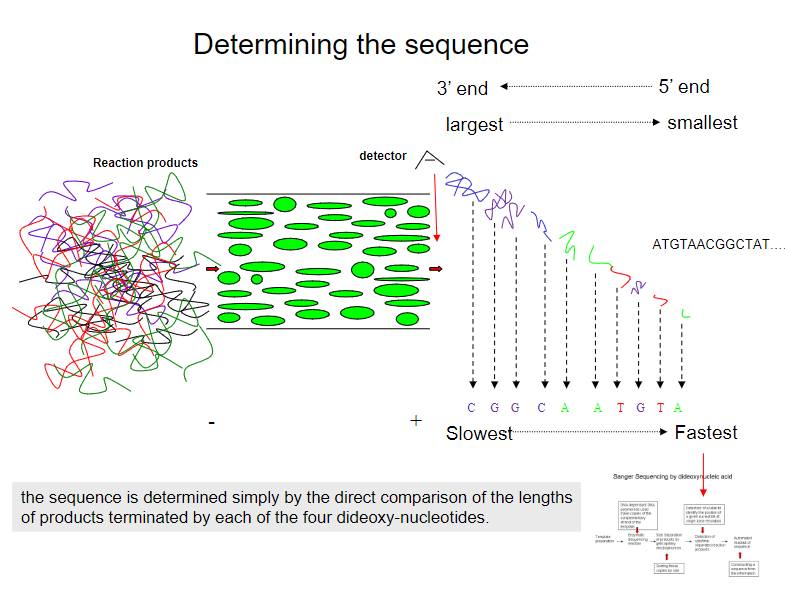
The sequence is determined by directly comparing the lengths of products terminated by each of the four dideoxy-nucleotides.
Each product corresponds to a specific nucleotide (A, T, C, or G) based on which dideoxy-nucleotide terminates the chain.
-
How is the base calling process automated in DNA sequencing using DNA-dependent DNA polymerase? (1)
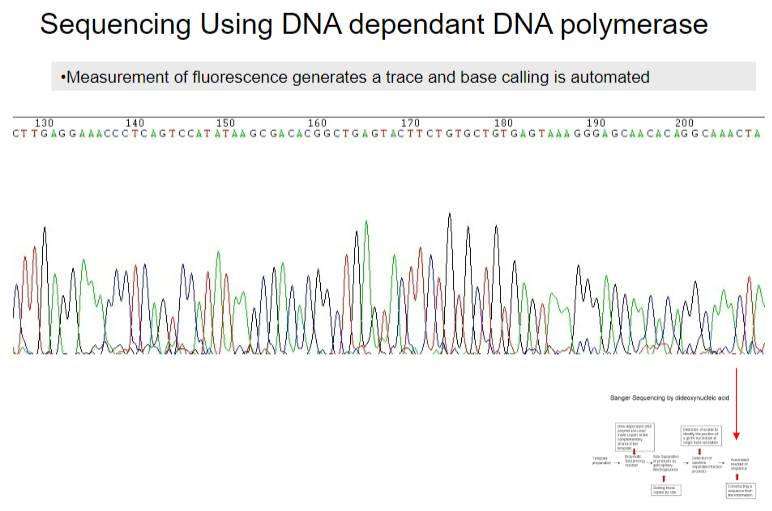
Base calling is automated through the measurement of fluorescence, which generates a trace.
-
What are some current applications of dideoxy sequencing in healthcare? (3)
Confirming specific genetic mutations in patients with suspected genetic diseases
Identifying mutations such as silent, missense, nonsense, truncating, indels, and mis-splicing
Identifying HIV haplotypes resistant to antiretrovirals for determining therapy (HAART)
-
What are some research applications of dideoxy sequencing? (4)
Sequencing mammalian and pathogen genes
Cloning or PCR amplicon sequencing to confirm a clone's sequence or site-directed mutagenesis
"Walking" a gene to identify a causative mutation in candidate gene studies
Confirming causative variants associated with genetic diseases following association studies
-
Summarize the key points of dideoxy sequencing. (5)
Introduced in the late 1970s and important in genomics
Involves two main components: the reaction and capillary electrophoresis
The reaction and approach are used to resolve DNA sequence
Applied as a confirmatory test in genetic mutations
Widely used in biomedicine, including pathogen gene sequencing and genetic disease studies

