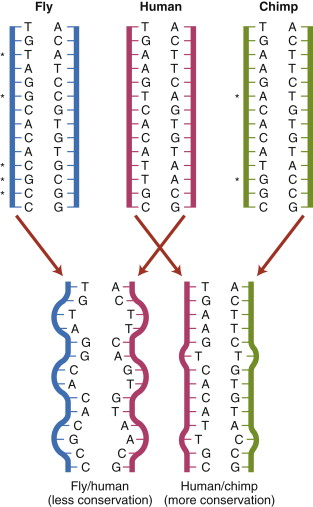
DNA and Hybridisation (Genomics)
SDL: DNA and Hybridisation DNA technology lies at the heart of modern genomics, diagnostics and significant areas of medical therapy. In addition, it is central to the developing area of personalised genetic based medicine, and important for our research and understanding of not only the human genome but that of model organism, pathogens, as well as many other environmental and commercially important organisms. This lecture will first revise the basic structure and properties and behaviour of nucleic acids. It will then discuss DNA strand dissociation (“melting”) and DNA strand annealing, in terms of both equilibrium and kinetics. It will introduce the concept that purified nucleic acids and short synthetic nucleotides that are complementary to specific sequences can anneal and form a hybrid double stranded molecule. This provides the theoretical basis for understanding nucleic acid hybridisation technologies, including Southern and northern blotting, Microarrays and is a pre-requisite for the formation of initiation complexes in molecular techniques for copying nucleic acids such as PCR and its role in diagnostics and DNA Sequencing including DNA Sequencing and Next Generation Sequencing all of which will be reviewed in subsequent sessions. Learning Outcomes On successful completion of the lecture, students should be able to: Describe the chemical and molecular make up and arrangement of DNA Explain how the structure and composition of DNA influences it chemical properties and behavior in solution Define and explain the concepts underpinning duplex formation, duplex stability, denaturation and renaturation Define the terms complementarity, specificity and stringency and explain factors influencing these Briefly describe the application of DNA hybridisation in some common probe based techniques used in Medical and Biomedical Sciences
-
What is the role of DNA technology in current research? (1)
DNA technology is central to current-day research, diagnostics, and therapeutics.
-
What are the key structural components of a nucleotide? (3)
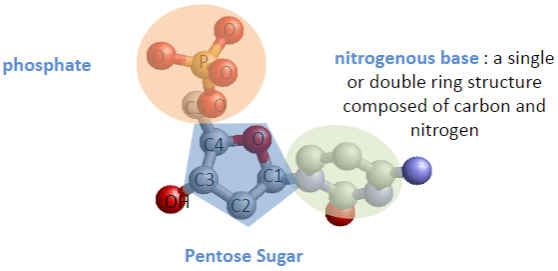
Nitrogenous base (single or double ring composed of carbon and nitrogen)
Phosphate group
Pentose sugar
-
Where do the nitrogenous base, phosphate group, and hydroxyl group attach on the pentose sugar? (3)
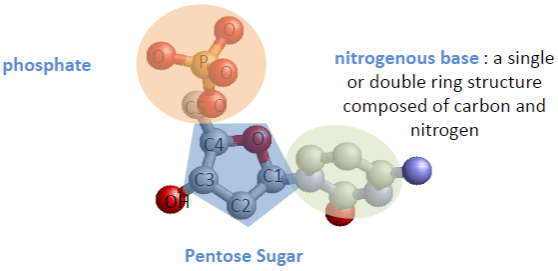
Nitrogenous base joins to carbon 1.
Phosphate group joins to carbon 5.
Hydroxyl group joins to carbon 3.
-
What are the four nucleotides in DNA? (1)
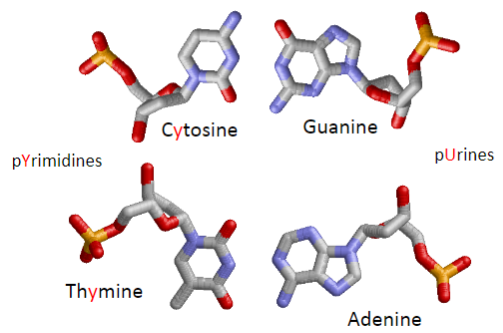
Cytosine, Guanine, Adenine, Thymine.
-
What is the difference between purines and pyrimidines? (1)
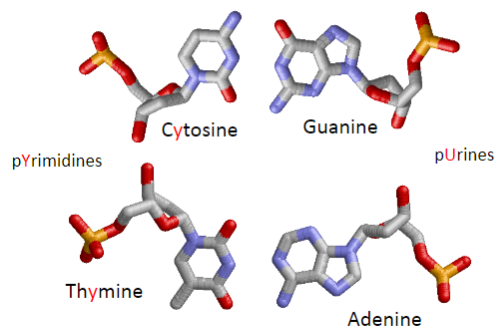
Purines (Adenine and Guanine) have two rings, and Pyrimidines (Cytosine and Thymine) have one ring.
-
What forms the basis of Watson and Crick base pairing? (1)
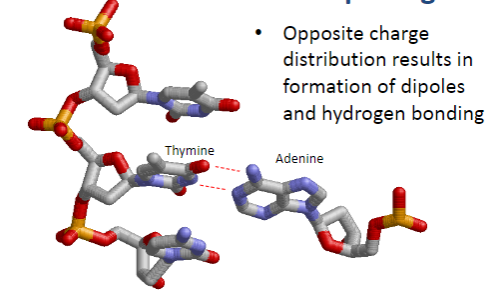
Hydrogen bonding due to the opposite charge distribution, resulting in dipoles.
-
How many hydrogen bonds form between base pairs? (1)
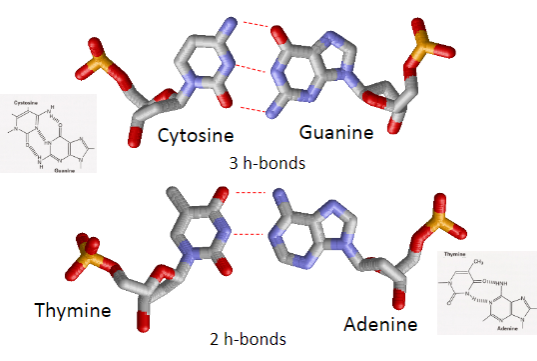
Cytosine and Guanine form 3 hydrogen bonds.
Adenine and Thymine form 2 hydrogen bonds.
-
What bonds link sugar phosphates in the DNA backbone? (1)
Phosphodiester bonds.
-
What stabilizes the arrangement of bases in the DNA double helix? (2)
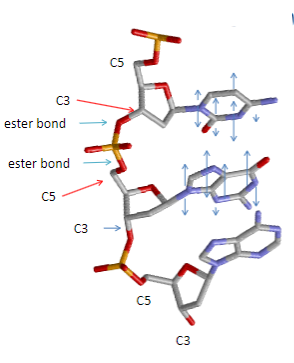
Base stacking (hydrophobic interactions) which arranges bases above each other and excludes water.
Van der Waals forces contribute to the stability.
-
What is the charge of double-stranded DNA and why? (1)
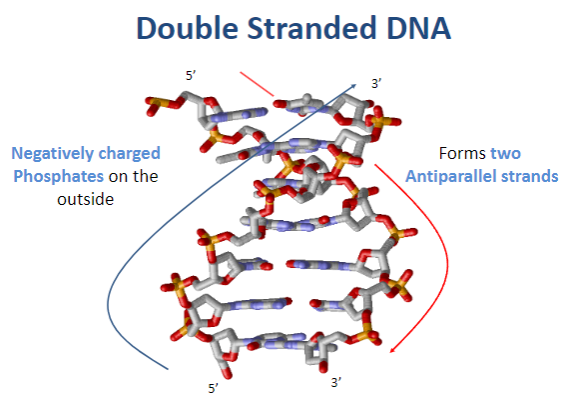
DNA is negatively charged due to the phosphates on the outside of the double helix.
-
How are the two strands of DNA oriented? (1)
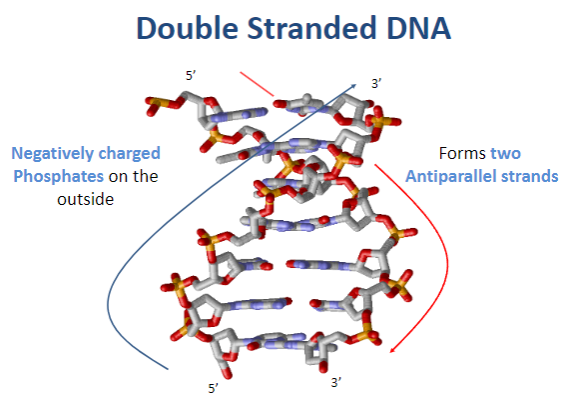
DNA strands are antiparallel.
-
What happens when DNA is denatured? (2)
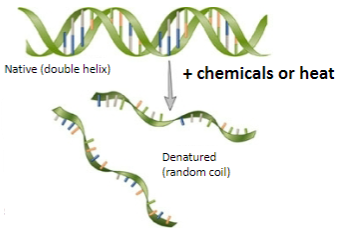
Double-stranded DNA converts to single-stranded molecules.
Hydrogen bonds within the helix are disrupted.
-
What factors can cause DNA denaturation? (2)
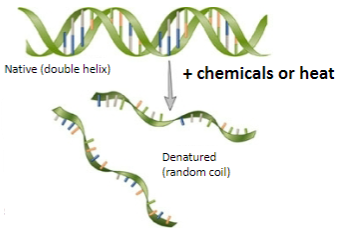
Heat
Strong alkali
Chemicals like urea.
-
How can DNA denaturation be measured? (2)
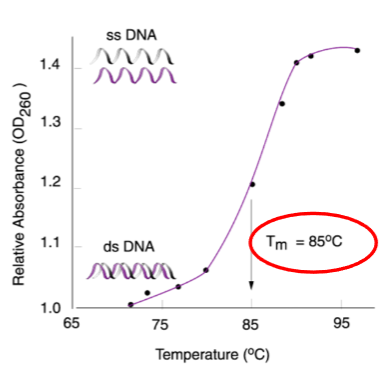
Denaturation can be measured optically by absorbance at 260nm.
The point at which 50% of DNA strands separate is called the melting temperature (Tm).
-
What is hyperchromicity? (1)
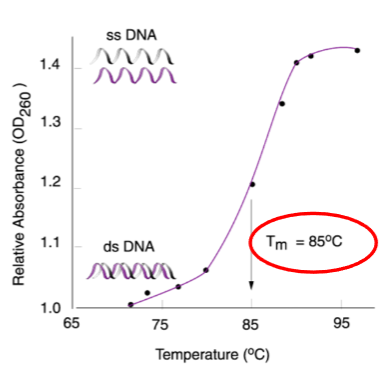
The increased absorption of light at 260nm during DNA denaturation.
-
What factors influence the melting temperature (Tm) of DNA? (5)
GC content.
Length of the DNA molecule.
Salt concentration
pH
Mismatches in base pairing.
-
How does GC content affect Tm? (1)
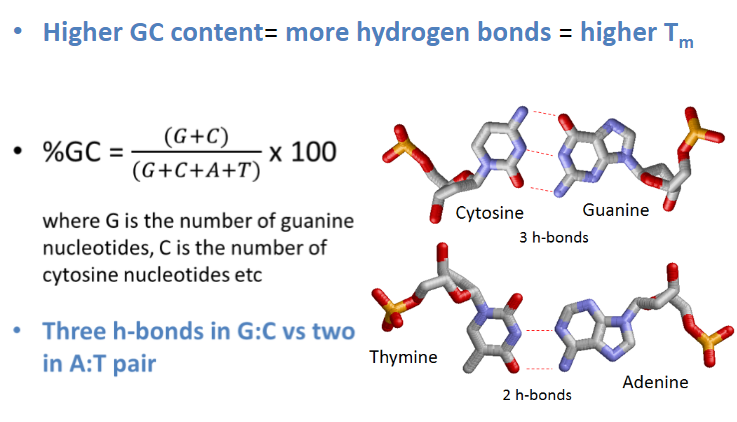
Higher GC content leads to more hydrogen bonds, resulting in a higher Tm.
-
How does the length of a DNA molecule affect Tm? (2)
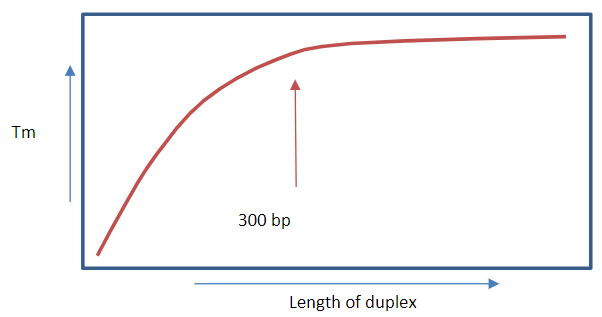
A longer contiguous duplex increases Tm due to more hydrogen bonds providing greater stability.
There is little further contribution to Tm beyond 300 base pairs.
-
How does salt concentration affect Tm? (1)
Salt stabilizes DNA duplexes.
Higher sodium ion concentration ([Na+]) results in a higher Tm.
-
What effect does increasing salt concentration have on mismatched base pairing? (2)
Increasing salt concentration stabilizes the DNA structure, increases Tm, and overcomes the destabilizing effect of mismatched base pairing.
This facilitates hybridization but decreases specificity.
-
How does high salt concentration affect base pairing specificity? (3)
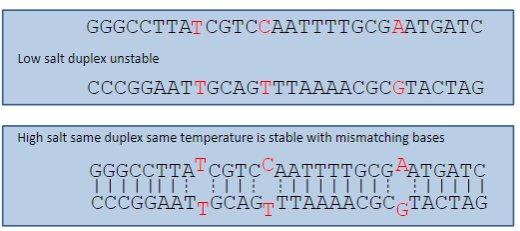
High salt concentration reduces the specificity of base pairing at a given temperature
-
What is the effect of chemical denaturants on Tm? (1)
Chemical denaturants disrupt hydrogen bonds, leading to a lower Tm.
Examples include alkali, formamide, and urea.
-
How does NaOH affect Tm? (2)
NaOH dissociates into Na⁺ and OH⁻, which disrupts hydrogen bond pairing.
Fewer hydrogen bonds result in a lower Tm.
-
What is the effect of high pH (alkalinity) on DNA duplexes? (1)
High pH destabilizes DNA duplexes, contributing to lower Tm.
-
What is a mismatch in DNA? (2)
A mismatch is defined as a base pair combination that is unable to form hydrogen bonds.
Mismatches reduce the number of hydrogen bonds, resulting in a lower Tm.
-
How do mismatches affect Tm? (3)
Mismatches lead to fewer hydrogen bonds, which decreases Tm.
Shorter contiguous stretches of double-stranded sequence also result in a lower Tm.
Mismatches distort the structure and destabilize adjacent base pairing.
-
What is denaturation in DNA, and is it reversible? (3)
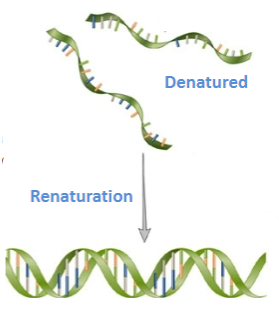
Denaturation is the process where double-stranded DNA separates into single strands.
It is reversible and referred to as renaturation.
-
What factors facilitate renaturation of DNA? (2)
Renaturation is facilitated by slow cooling and neutralization.
The formation of the structure favors energy minimization driven by the change in free energy (ΔG)
-
What is Renaturation? (1)
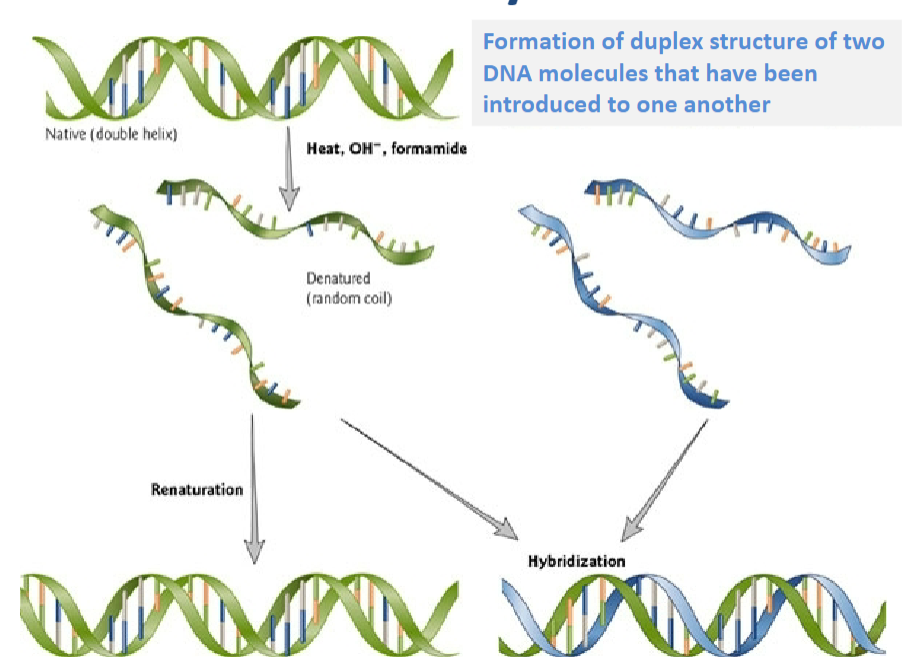
Renaturation is the process of reforming a duplex structure from two DNA molecules that have been introduced to one another.
-
What is Hybridisation? (2)
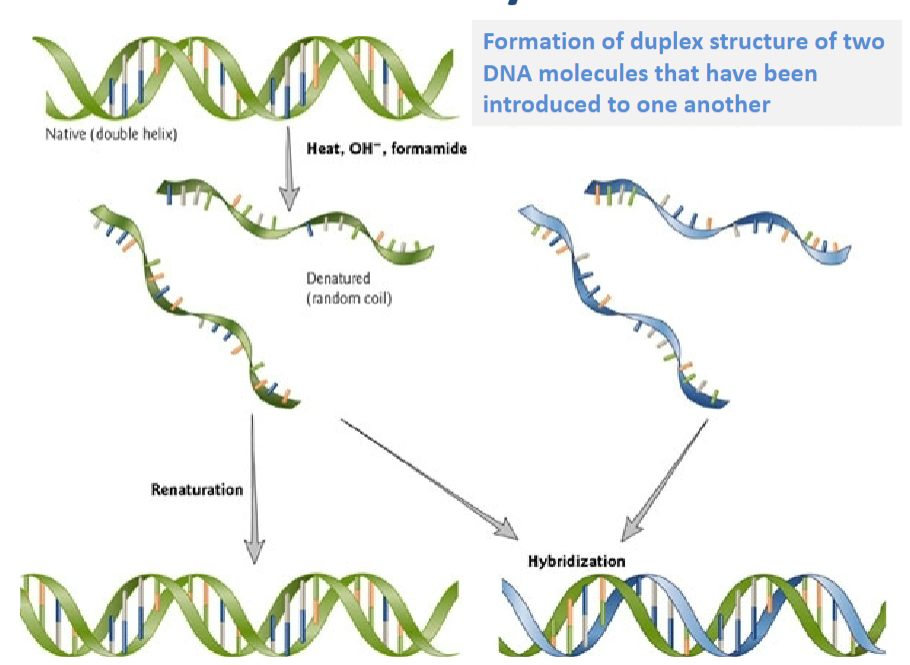
Hybridisation refers to the formation of a duplex structure between complementary DNA or RNA strands.
-
What is the significance of complementarity and Tm in DNA hybridisation? (1)

Complementarity and melting temperature (Tm) determine the specificity of DNA hybridisation; mismatches reduce Tm, while perfect matches are thermodynamically favoured.
-
How can we use the property of Tm in DNA hybridisation? (1)

The property of Tm can be used to form a complementary molecule with no mismatches, ensuring higher specificity.
-
What is stringency in the context of hybridisation? (1)
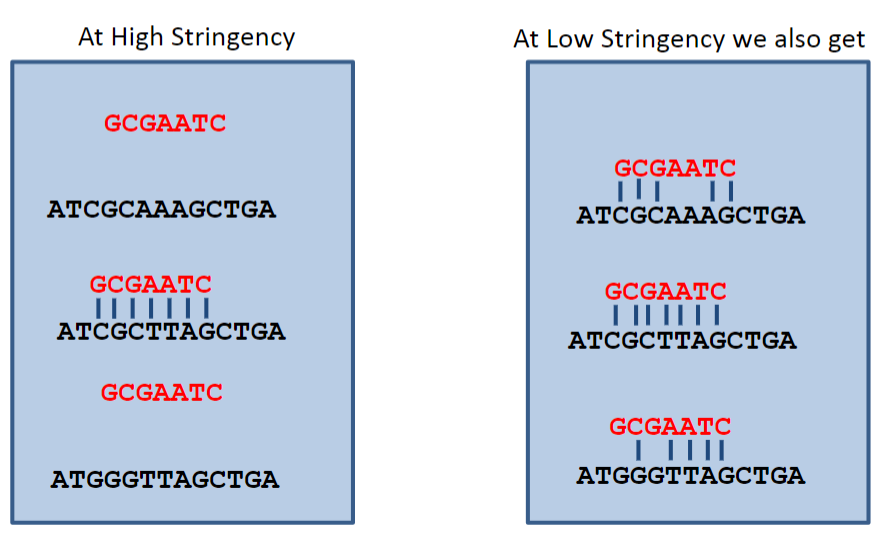
Stringency refers to manipulating conditions to limit hybridisation between imperfectly matched sequences, allowing for increased specificity in DNA interactions.
-
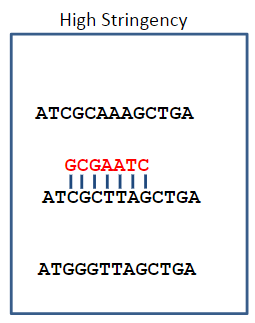
What happens under high stringency conditions in DNA hybridisation? (1)
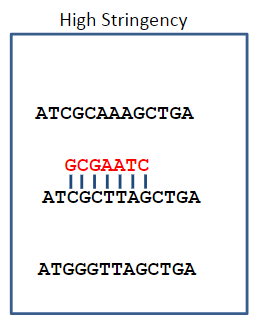
Under high stringency, only complementary sequences are stable, which is determined by a temperature near Tm or a low salt concentration.
-
How does manipulating conditions affect hybridisation specificity? (1)
Manipulating conditions to limit hybridisation between imperfectly matched sequences increases specificity in DNA interactions.
-
How can we measure duplex formation? (1)
Duplex formation can be measured, and the melting temperature (Tm) of a duplex can be determined.
-
How can duplex formation be manipulated? (1)
Duplex formation can be manipulated by controlling factors such as high and low stringency conditions.
-
What techniques rely on complementarity and hybridisation? (6)
Techniques relying on complementarity and hybridisation include:
Chromosome Painting
Microarrays
Exon Capture
DNA Sequencing (Sanger or Next Generation Sequencing)
PCR and its various forms
Recombinant DNA techniques like cloning.
-
What is the purpose of nucleic acid hybridisation techniques? (1)
Nucleic acid hybridisation techniques identify the presence of nucleic acids containing a specific sequence of bases, allowing for the capture and quantitation of these sequences in a mixture.
-
What is hybridisation? (1)
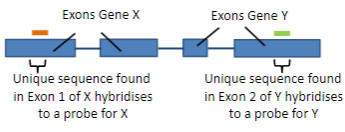
Hybridisation uses the ability of nucleic acids to form specific duplexes through the complementarity of sequences.
-
What are probes in the context of hybridisation? (1)
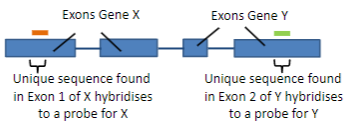
Probes are labelled nucleic acids used in hybridisation to identify specific sequences by forming duplexes with complementary target sequences.
-
What is a probe? (1)
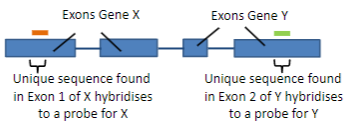
A probe is a single-stranded DNA (ssDNA) or RNA molecule, typically 20 to 1000 bases in length.
-
How is a probe labelled? (1)
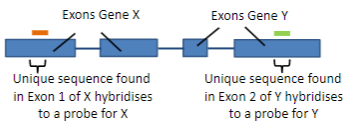
A probe is labelled with a fluorescent or luminescent molecule, and less commonly with a radioactive isotope.
-
How many probes can be used simultaneously in some techniques? (1)
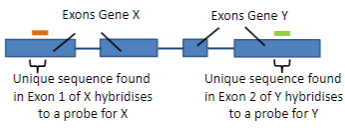
In some techniques, thousands or millions of probes can be used simultaneously.
-
What are common targeted hybridisation-based techniques? (1)
Common targeted hybridisation-based techniques include in-situ hybridisation of tissue sections, chromosome painting of chromosome spreads, analysis of mRNA or DNA by PCR/qPCR, and Sanger/dideoxy sequencing.
-
What is a limitation of common targeted hybridisation-based techniques? (1)
These techniques are not very scalable and only detect a few genes at a time and small numbers of samples.
-
What are genome-wide techniques used for? (1)
Genome-wide techniques are used for measuring gene expression or genomic composition.
-
What high-throughput techniques employ hybridisation? (1)
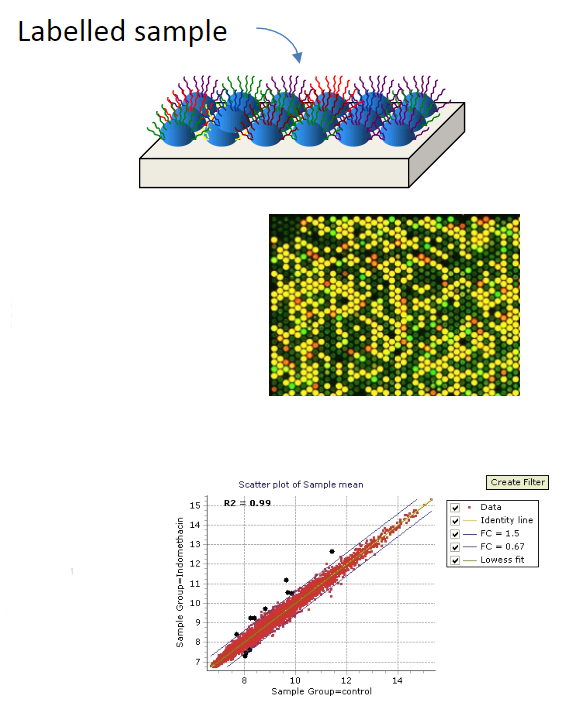
High-throughput techniques that employ hybridisation include microarrays, sequence capture (e.g., exon capture), and next-generation sequencing.
-
What are microarrays? (1)
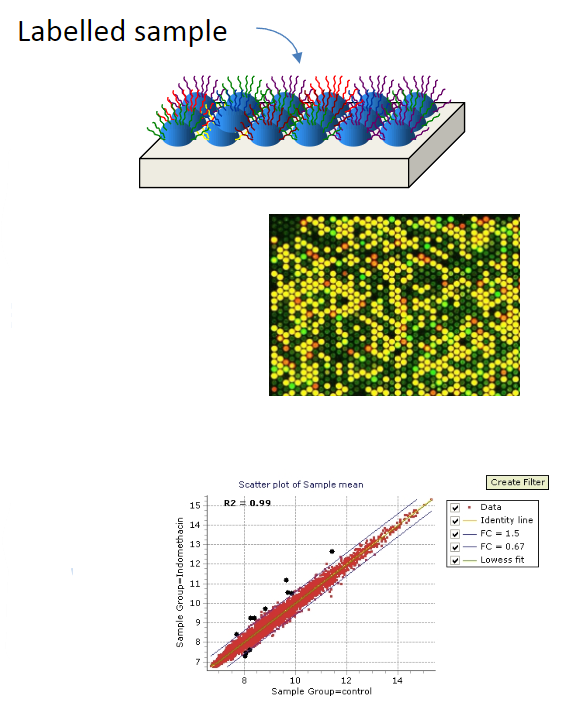
Microarrays are an ordered assembly of thousands of nucleic acid probes fixed to a solid surface, which are hybridised with a sample of interest
-
What is next-generation sequencing? (1)
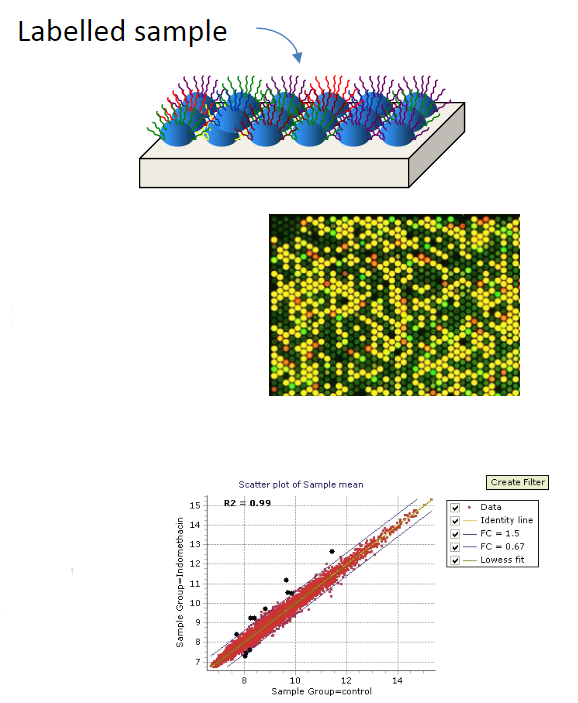
Next-generation sequencing is the parallel sequencing of millions of molecules captured on a surface by hybridisation.
-
What is the summary of hybridisation techniques? (1)
Hybridisation techniques involve the use of probes, which allow for various applications, including chromosome painting and qPCR, and high-throughput techniques like microarrays and next-generation sequencing.

