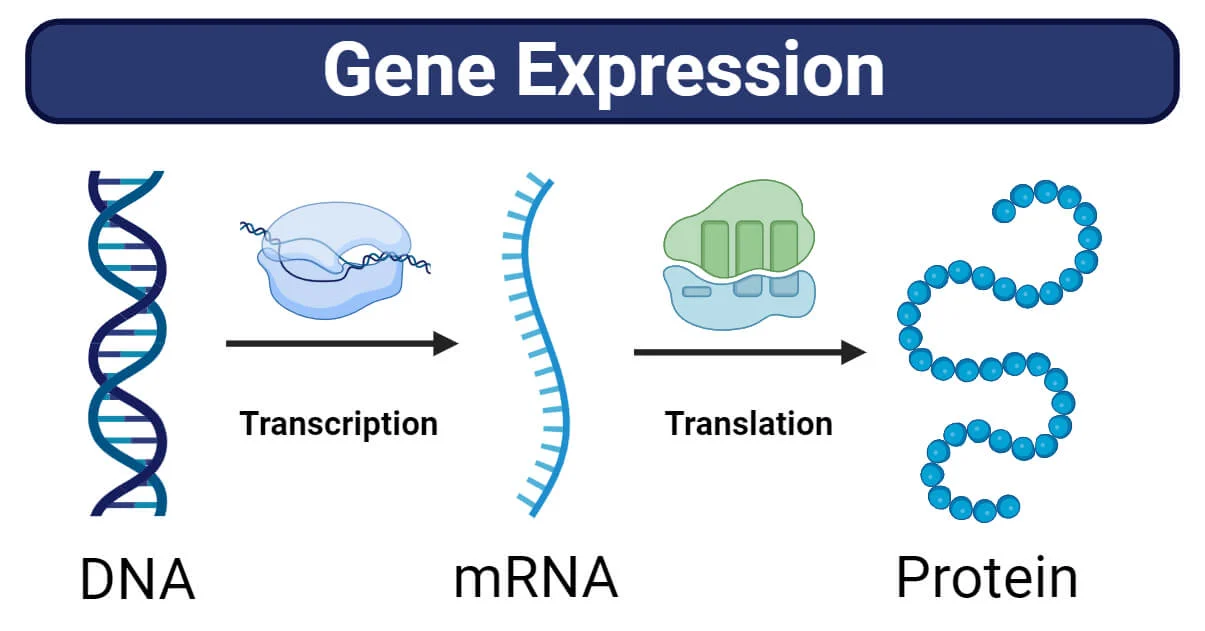
Control of gene expression (The Living cell)
Learning Outcomes At the end of the session students will be able to Describe what is meant by genotype. Understand the importance of differential gene expression. Describe what is meant by transcriptome Describe the anatomy of promoters Explain the recruitment of RNA polymerases to bacterial and eukaryotic promoters Describe what is meant by consensus DNA sequences Distinguish general transcription factors from regulatory transcription factors Describe transcriptional switches in prokaryotic and eukaryotic cells Appreciate how our understanding of transcription regulation has resulted in the development of anti cancer drugs Describe how deregulated gene expression results in disease. Understand what is meant by post-transcriptional gene regulation Describe mechanisms of post-transcriptional gene regulation Describe microRNAs and their mechanism of action
-
What is the genotype?
Definition: The genetic blueprint of an organism, representing its complete set of genes.
Key Point: All human cells share the same genotype.
-
Facts about protein coding genes: How many protein coding genes are there in the human genome? And how many of these genes are expressed?
Genetic Estimate: The human genome is estimated to contain 20,000 protein-coding genes.
Cell Expression: Despite the total number, only about 12,000 genes are expressed in any given cell type.
-
What is differential gene expression?
Concept: The process by which genes are selectively turned on or off in different cells at different times.
-
Why is differential gene expression important?
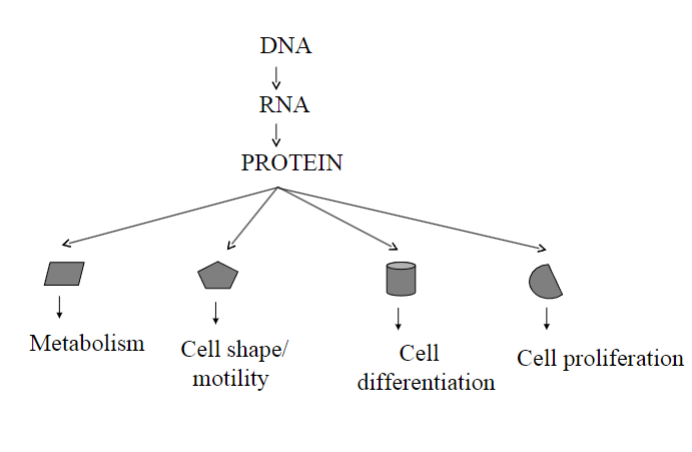
Differential gene expression is crucial because it allows cells to specialize and perform specific functions within an organism by selectively activating or deactivating genes.
-
Failure to regulate gene expression tightly may lead to what?
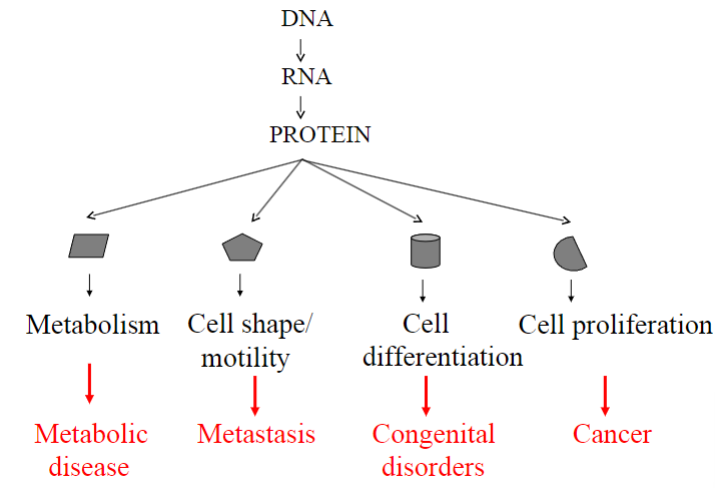
-
State some information about the transcriptome:
•The complete set of RNA transcribed from a genome
• Most of the eukaryotic genome is never transcribed; only about 50% of the prokaryotic genome is transcribed
-
Picture outlining the different levels of gene transcription
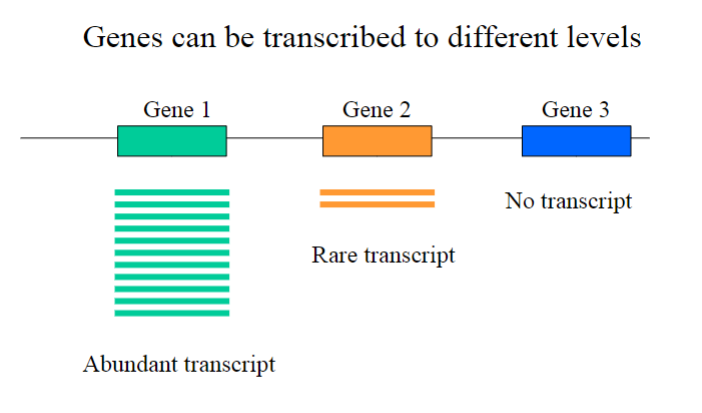
Some genes might be tissue specific
-
What is a housekeeping gene? (4)
Housekeeping Gene:
-Continuously expressed in all cells.
-Essential for basic cellular functions.
-Provides a baseline for comparing gene expression.
-Often used as internal controls in gene expression studies.
-
What is an inducible gene?
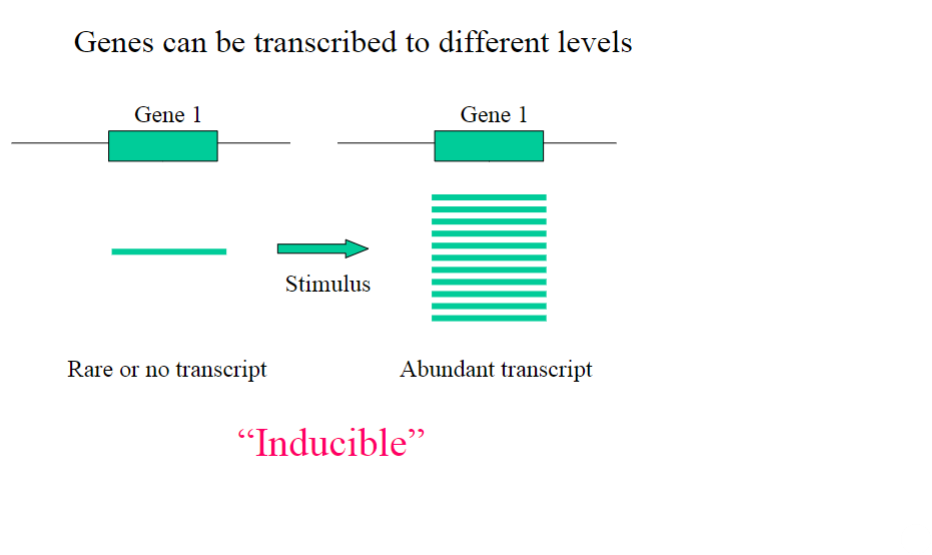
Inducible Gene:
Activated or upregulated in response to specific stimuli or environmental conditions.
-
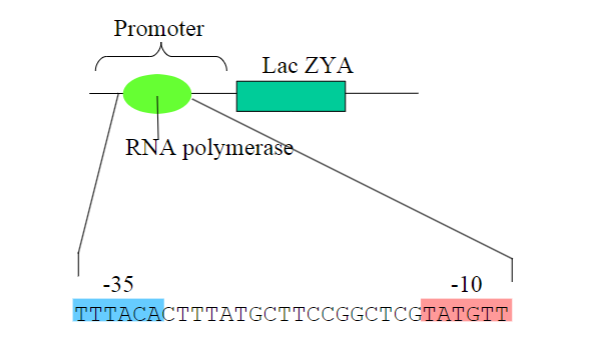
The sequence immediately 5’ to the region to be transcribed is called a what?
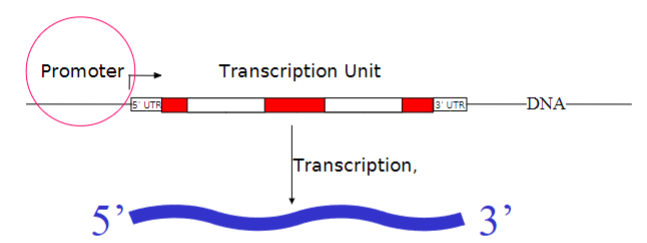
Promoter region
-
Promoters will recruit what to a DNA Template?
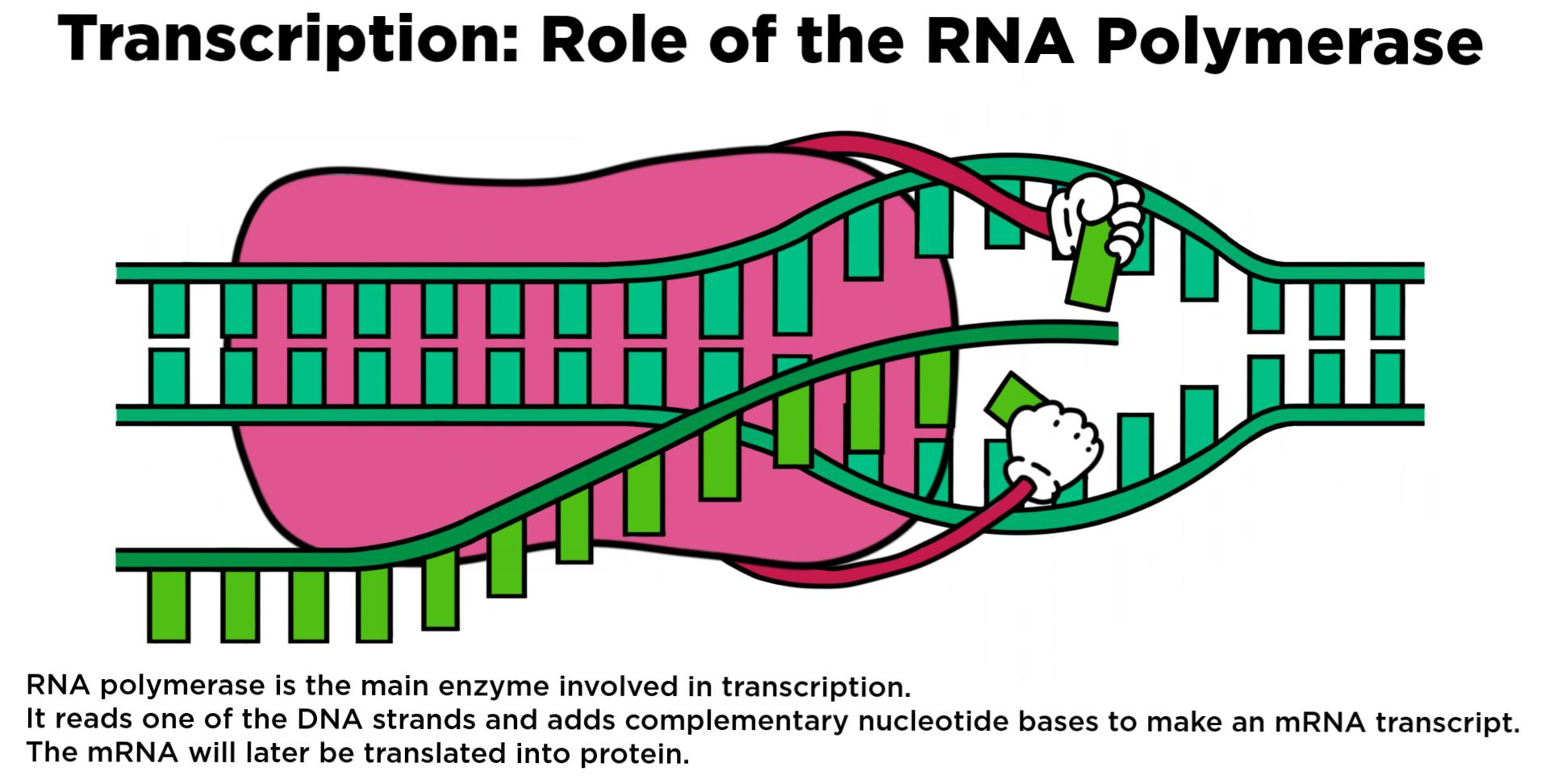
RNA polymerase
-
Recruitment of the RNA polymerase via the promoter cannot be done alone; Recognition of promoters is mediated by initiation factors… What are they? (for both prokaryotes and eukaryotes)
– Predominantly “Sigma factor” for prokaryotes– The TFII basal transcriptional machinery for eukaryotes
• In both prokaryotes and eukaryotes, they function to dramatically alter the level of recruitment of RNA polymerase and/or its ability to initiate transcription
• Additionally, in eukaryotes they can influence local chromatin structure
-
What is the consensus sequence?
Consensus Sequence:
Represents the most common nucleotide or amino acid at each position in a sequence alignment.
-
The ability of sigma factor and TFII to recruit RNA polymerases to promoters are GENERIC: what does this mean?
They happen at every promoter
-
What are the Transcriptional switches (both prokaryotic and eukaryotic)
• Prokaryotic– The lac operon
• Eukaryotic– Transcription factors and regulatory elements
-
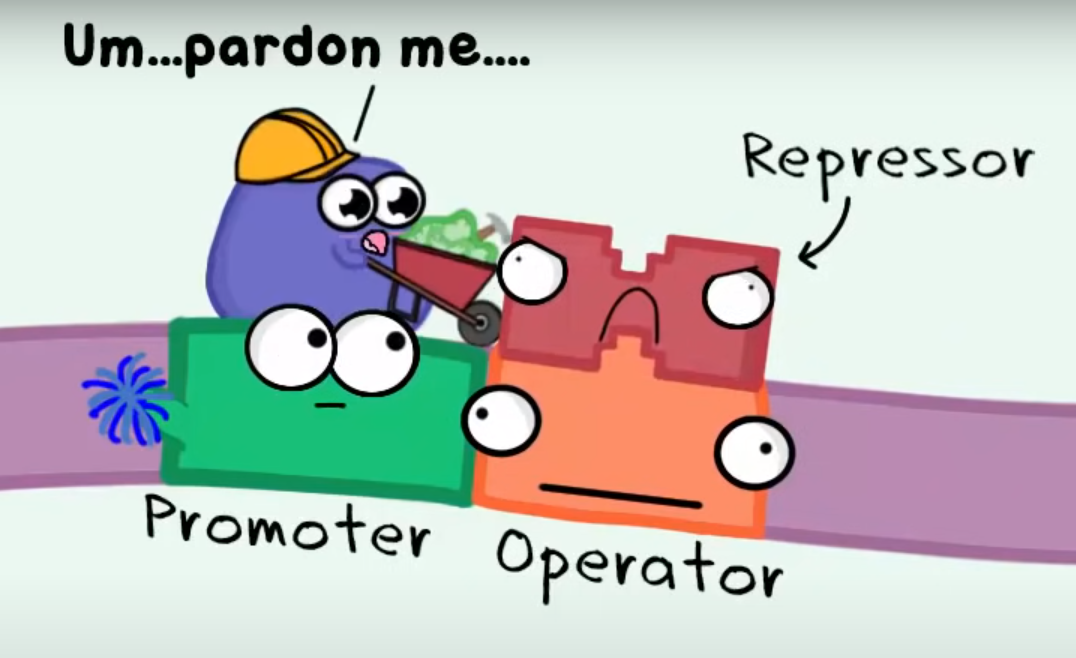
What is the Lac repressor?
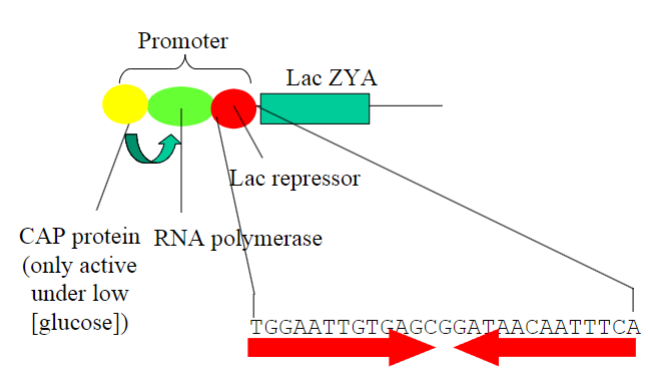
Lac Repressor:
-Regulatory protein in bacteria, notably in E. coli. (prokaryotes)
-Controls the lac operon, which comprises lacZ, lacY, and lacA genes.
-The lac repressor is a protein that represses (inhibits) transcription of the lac operon
-Genes encode enzymes for lactose metabolism.
-
The molecular basis of transcriptional control is well understood in prokaryotes; when is the CAP protein active in transcription?
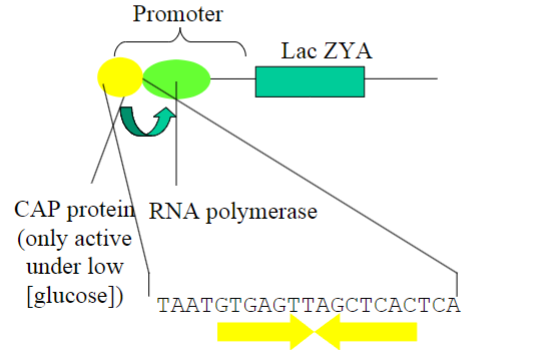
Under low glucose
-
What are the triggers and corresponding transcription factors? (do not state the regulatory elements)
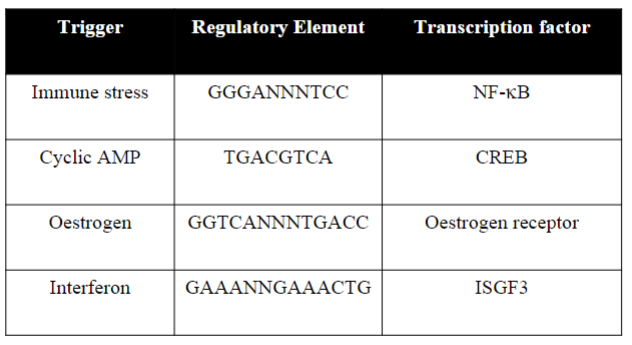
-
Transcription is regulated by?
....the efficiency of recruitment of RNA polymerase II and Regulatory factors
-
What do regulatory transcription factors do?
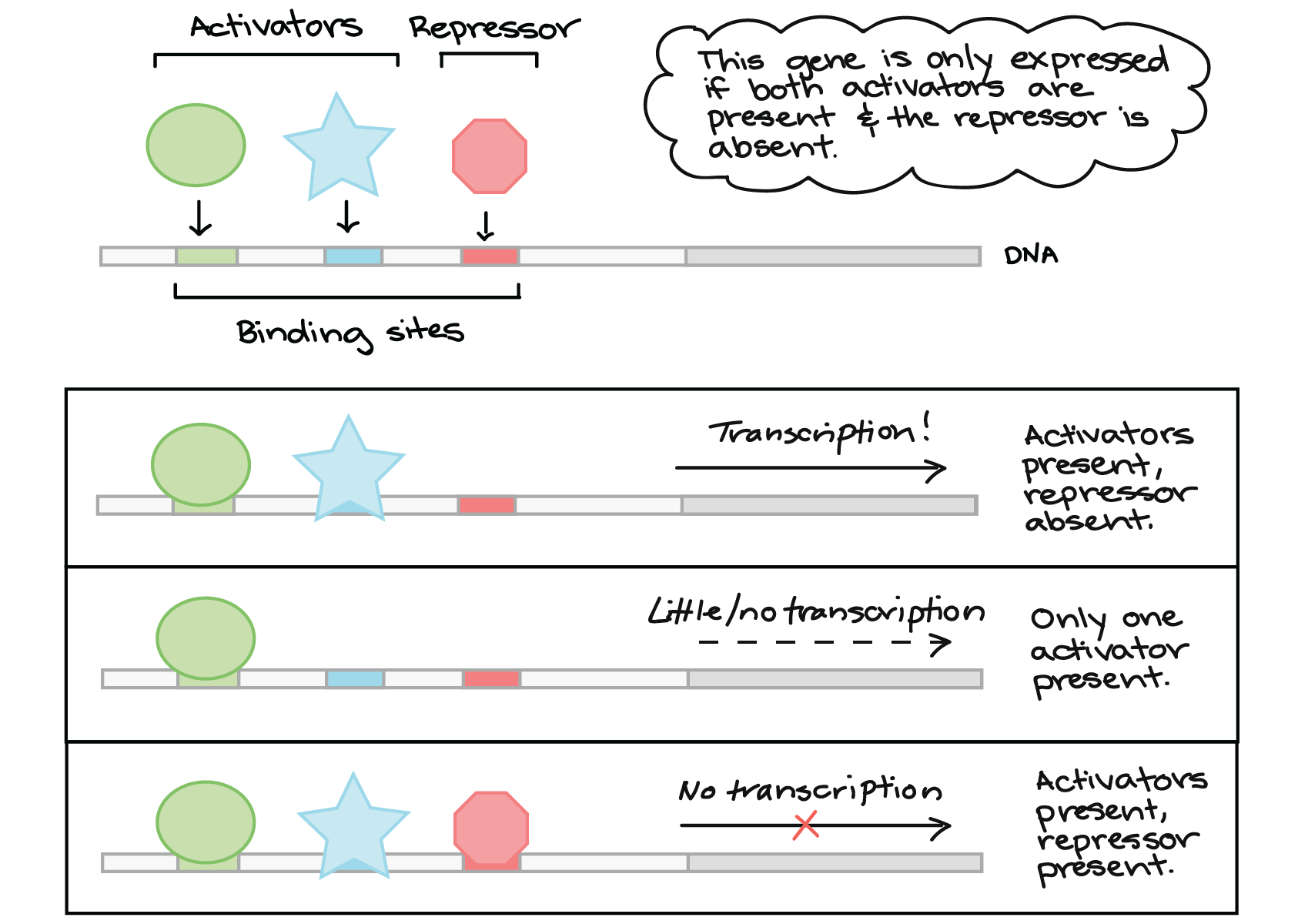
-Regulatory transcription factors bind to the promoter and help recruit general transcription factors
-
What Is one way of interfering with the interactions between regulatory and general transcription factors?
Therapeutic drugs
-
What are Ubiquitous factors?
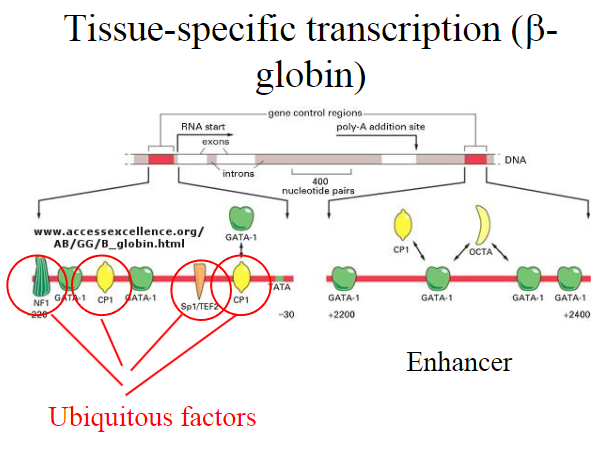
Ubiquitous Transcription Factors:
Proteins that play a role in the regulation of gene expression.
Often bind to DNA promoter regions to initiate or facilitate transcription.
-
What are tissue specific factors?
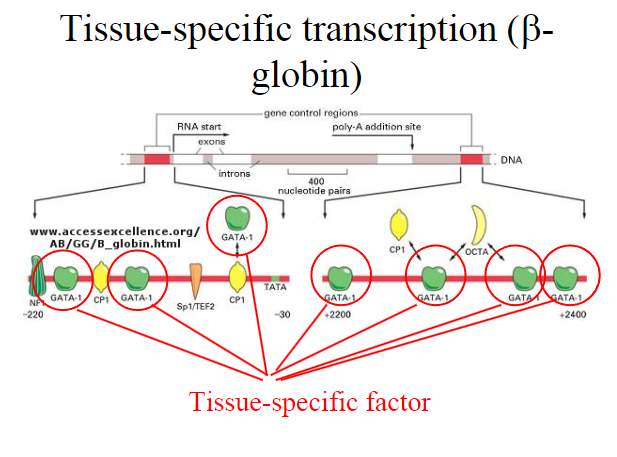
Tissue Specific Factors:
Tissue-specific factors are expressed in specific tissues or cell types.
Their expression is often regulated in a manner that restricts their presence to particular developmental stages or physiological conditions.
-
What is the restriction point in the cell cycle?
It Is A specific checkpoint in late G1 phase, where a cell becomes committed to progressing through the cell cycle and entering the S phase for DNA synthesis.
-
What is the mitogenic signal in the cell cycle?
The mitogenic signal is a signalling event that stimulates a cell to enter the cell cycle and progress through the G1 phase, ultimately leading to cell division.
-
Transitions through the cell cycle are regulated by what? Elaborate
Cyclin dependent kinases!
•They play a key role in the event of G1/S transition which is the transcriptional activation of genes
•CDKs target transcription factors
-
What is the factor that activated the promoters for G1/S transition genes?
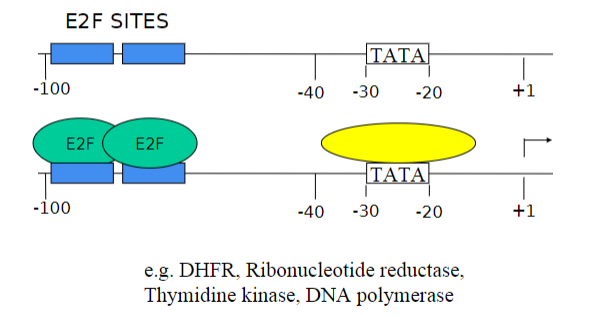
E2F
-
Can you describe the activity of E2F?
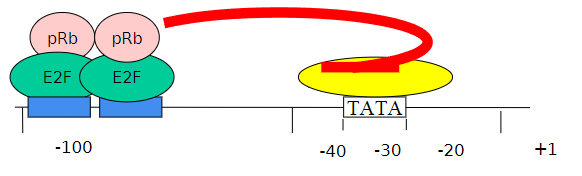
E2F activity is repressed in G0 and early G1 by the product of the Retinoblastoma gene (pRB)
-
What is the G0 phase of the cell cycle?
G0 is a quiescent state where cells temporarily or permanently exit the cell cycle and cease active proliferation.
-
How does mitogenic stimuli effect E2F-pRB activity?
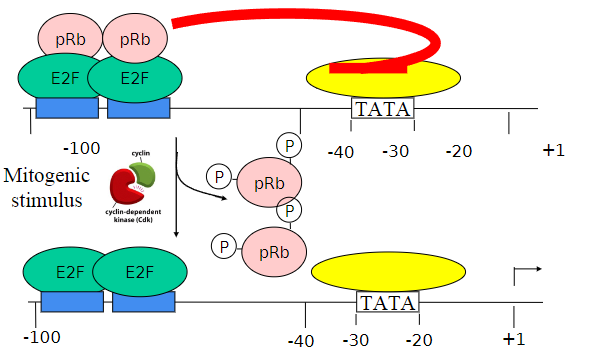
It will activate the Cdks, and pRB proteins are phosphorylated so it can no longer bind to the E2F molecules
-
Rb (Retinoblastoma) is a common target in what?
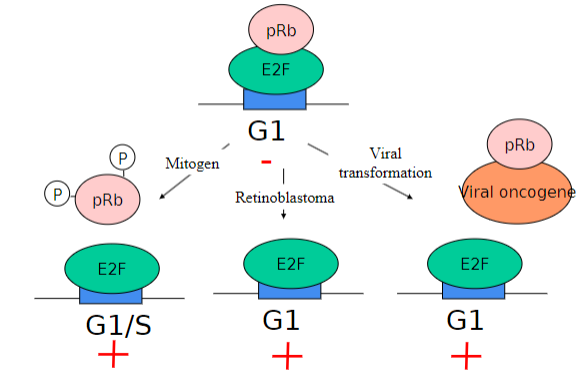
CANCER!!!!
-
What are enhancers? (3)
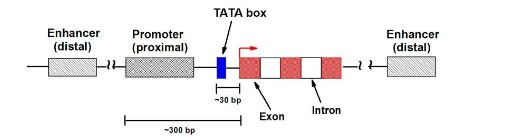
• Sequences of DNA that are not immediately adjacent (next) to where transcription starts that act to enhance the recruitment of RNA polymerase to a promoter
• Enhancers can reside 5’ or 3’ end to a transcription unit, and can even be located within an intron
• Like promoter enhancers contain DNA sequences that are very strong binding sites for specificity factors or “transcription factors”
-
What are the 3 examples (1 specific) of translational control? Elaborate on them
• Early embryogenesis – during the first 4 – 8 cell divisions, there is virtually no gene expression. At the end of blastocyst formation, the first genes to be expressed are due to up-regulation of translation from maternally-derived pre-formed mRNAs
• Environmental stress – exposure to heat shock or pathogens can cause global changes in translation
• Many specific examples – e.g. ferritin
-
Does the 5' UTR determines whether the ribosome binds to the mRNA strand?
NO
but..... It DOES play a major role in determining how efficiently the ribosome initiates translation
For example:– Globin (very efficiently translated)– Ferritin (very inefficiently translated)
-
STATEMENT: Intracellular Iron levels are translationally controlled; Elaborate on this
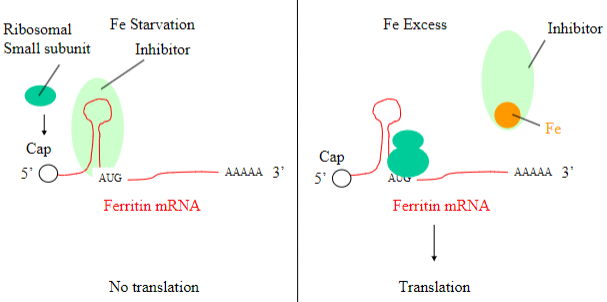
•Ferritin - binds Iron and retains in the cytoplasm as a store for excess
•Only need Ferritin at times of Iron excess
-
3’ UTR can play a role in determining the stability of....what? Elaborate
The mRNA
– Globin 3’UTRs confer stability
– For example: Immune stress hormones are produced from very unstable mRNAs
-
What are small. Non-coding RNAs/miRNAs
• The human genome encodes over 2000 small non-coding RNAs that are transcribed by RNA polymerase II
• These RNAs are referred to collectively as microRNAs or miRNAs
• These miRNAs act to control the post- transcriptional regulation of as many as one-third of all human genes!
• Any given miRNA can regulate several target genes
-
Picture outlining how Micro RNAs are derived by processing from a larger precursor
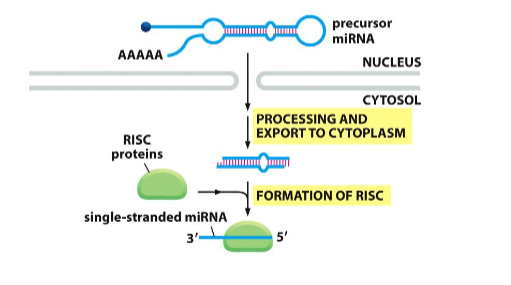
-
Picture outlining the Mechanism of action of miRNAs
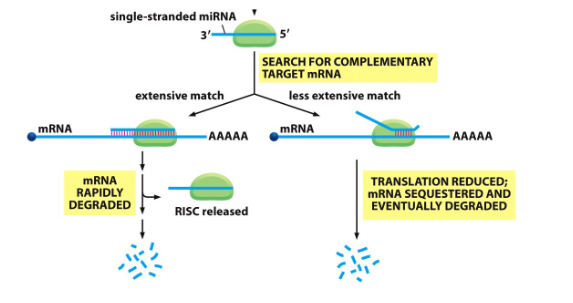
-
What is b-thalassaemia?
• A group of genetic diseases caused by insufficient expression of b-globin
• In most types of b-thalassaemia, the b-globin protein is structurally normal (unlike sickle cell disease)
• There are Multiple independently arising forms of the disease
-
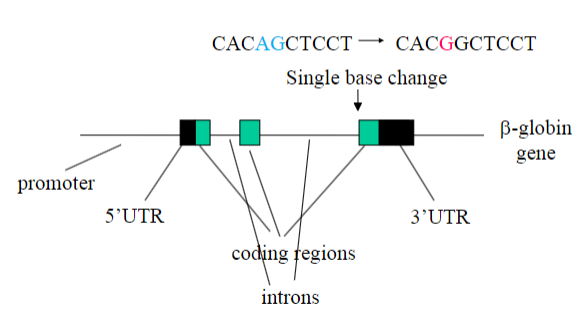
Picture outlining how Mutations in b-thalassaemia map/localise to multiple sites in the b-globin gene:
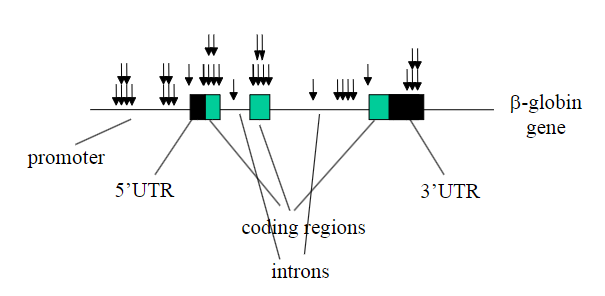
Arrows represent point mutations which stop the genes being expressed

