Chromatin Structure and Histone Code (The Living Cell)
Desired Learning Outcomes By the end of this lecture, students should be able to: · Understand the main functions and properties of the chromatin. · Explain the structure of a nucleosome. · Describe the different levels of DNA organisation in the nucleus: from nucleosome to chromosome. · Understand the main mechanisms of chromatin remodelling –histones modifications and histone code.
-
What is the definition of Chromatin?
-Chromatin: nuclear complex of DNA and associated proteins that forms chromosomes within the nucleus of eukaryotic cells.
-
What is the function of Chromatin?
- Function: Packaging long DNA molecules into more compact and denser shape (small volume) to fit into the nucleus and to protect the DNA structure and sequence
-
Who discovered the DNA?
Rosalind Franklin (1952. King's College London)
-
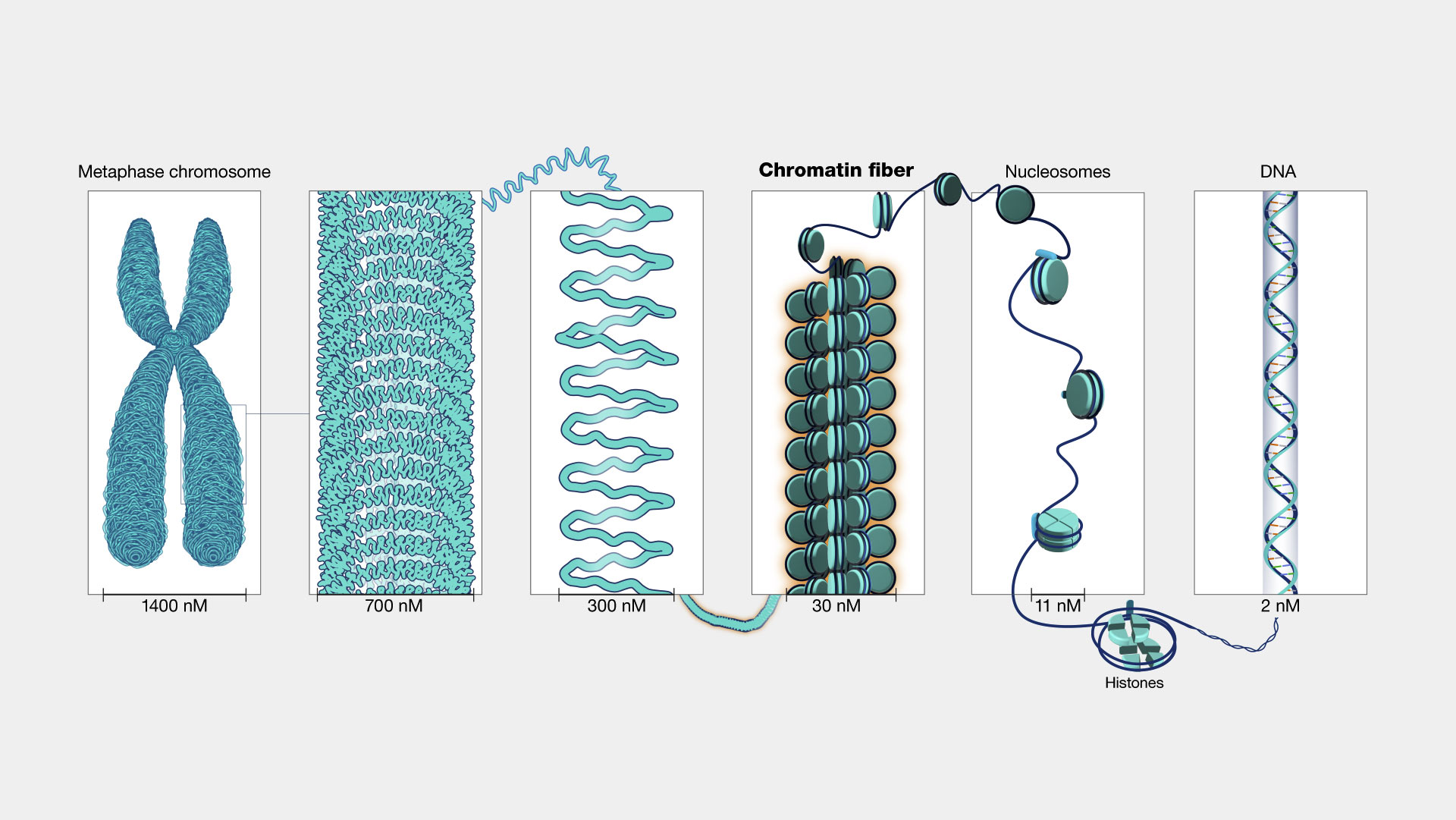
Outline the four levels of DNA packaging:
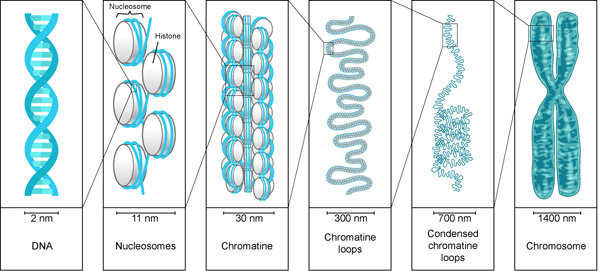
1st Level:
- The organisation of DNA and histones into nucleosomes- Increases DNA packaging 7-fold- ≈10nm
2nd Level:- Nucleosomes pack themselves in fibres- Increases DNA packaging 6-fold- 30nm
3rd Level:- Fibres pack themselves into loops and TADs (Topologically associating domains) and form chromatin-Increases DNA packaging 3-fold- 100-250nm
4th Level:- Represented by the mitotic chromosome-Means 1000-fold packaging
- 700-1000nm
-
In interphase, chromatin is organised in .... what?
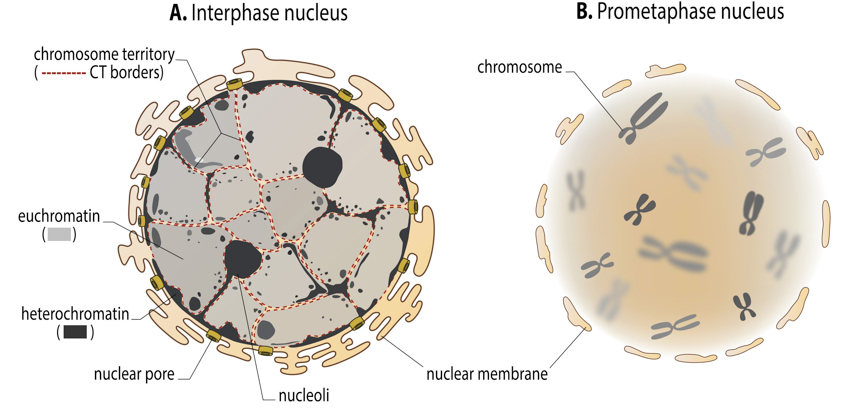
- In interphase, chromatin is organised in euchromatin and heterochromatin
-

State some facts about Euchromatin: (3)
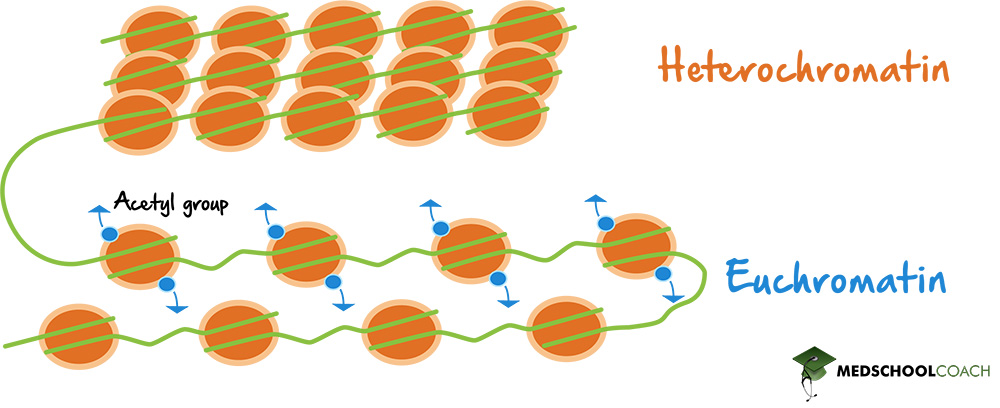
• Low density
•Lightly staining areas of chromatin
• Rich in genes.
-

State some facts about Heterochromatin: (4)
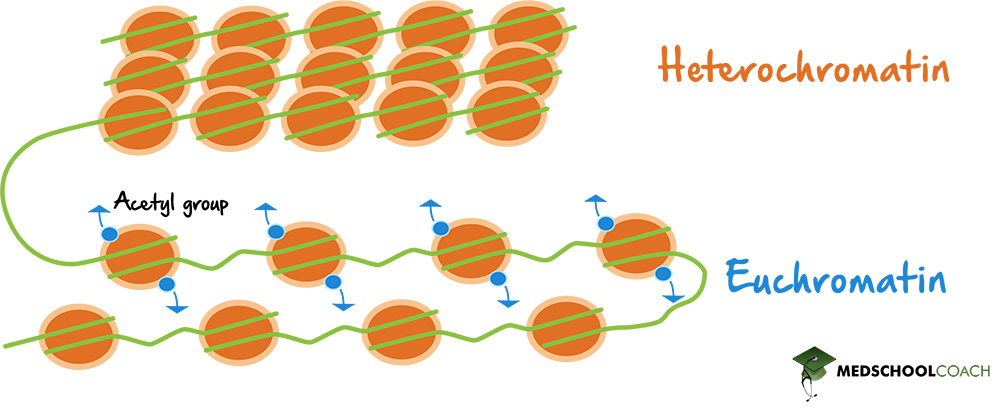
• Highly condensed, 30nm fibre.
• Darkly staining areas of chromatin, often associated with nuclear envelope.
• Gene poor
-
What is Constitutive heterochromatin?
-Constitutive (genes that are transcribed in an ongoing manner)
Constitutive heterochromatin remains in a condensed (inactive) state throughout the cell cycle.
It does not switch between euchromatic and heterochromatic states.
Typically not involved in active transcription.
Maintains a stable structure and is not as dynamic as facultative heterochromatin.
-
What is Facultative heterochromatin?
-Facultative (occurring optionally in response to circumstances rather than by nature)
Facultative heterochromatin can switch between euchromatic (active) and heterochromatic (inactive) states.
It is not fixed and undergoes changes during cellular development or in response to environmental signals.
Changes involve modifications to histones and DNA methylation.
Plays a role in regulating gene expression based on cellular needs
-
Chromatin shows plasticity: enabled by what?
- Chromatin shows plasticity: enabled by choice of histone variants, modifications of DNA bases, and reversible post-translational modifications (PTM) of histone tails.
-
What is the Nucleosome?
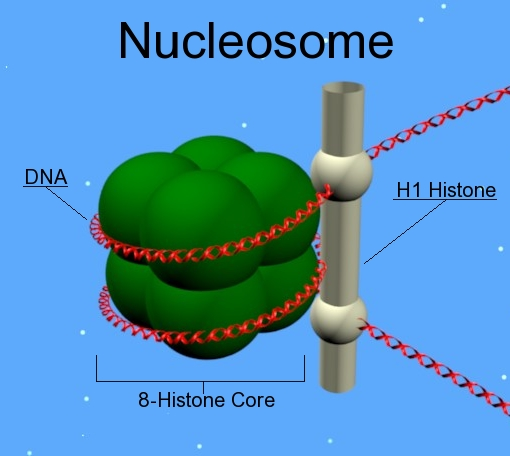
- Nucleosome: fundamental structural unit of chromatin. Composed of a little DNA wrapped around proteins called histones.
-
What do histones assemble to form? Elaborate (2)
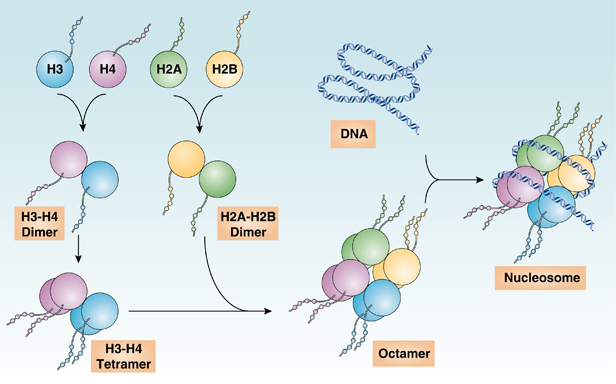
-Histones assemble to form an octamer core:
• H2A, H2B, H3 and H4 (2 molecules each)
• N-terminal tails stay outside the octamer core
-
What do the Nucleosomes consist of? (4)
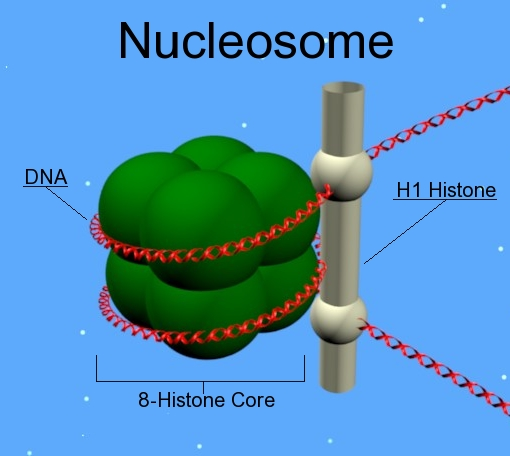
- 146bp of DNA wrapped around the histones core (1.7 turns of DNA).
- H1 protein works like a clip and keeps DNA wrapped around nucleosome
- Linker DNA 38-53bp DNA that interconnects two adjacent nucleosomes. It helps to stabilize the zigzagged 30 nm chromatin fibre. Gives the appearance of 'beads on a string'
-Nucleosomes then tightly pack together into a fibre of 30nm
-
Nucleosomes dynamics: during transcription, or DNA replication, nucleosomes must be what?
They must be removed from the DNA in front of the polymerase, and replaced behind the polymerase.
-
When do Chromatin loops occur? Also what is Cohesin?
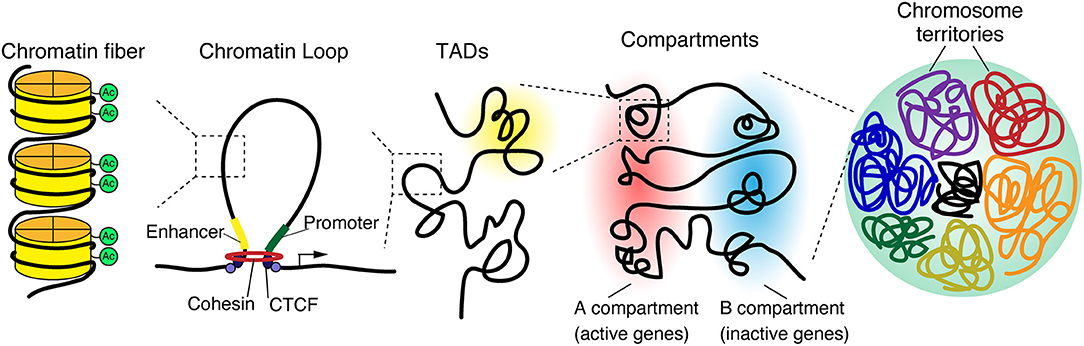
-Chromatin loop formation (looping): occurs when stretches of genomic sequence that reside on the same chromosome are in close physical proximity to each other.
-Cohesin: protein ring that binds to DNA and facilitates loops
-
What is a positive effect of hairpin loops?
Chromatin loops provide favourable environment to processes such as DNA replication, transcription, and repair.
-
What are TADS an abbreviation of?
Topologically Associating Domains (TADs)
-
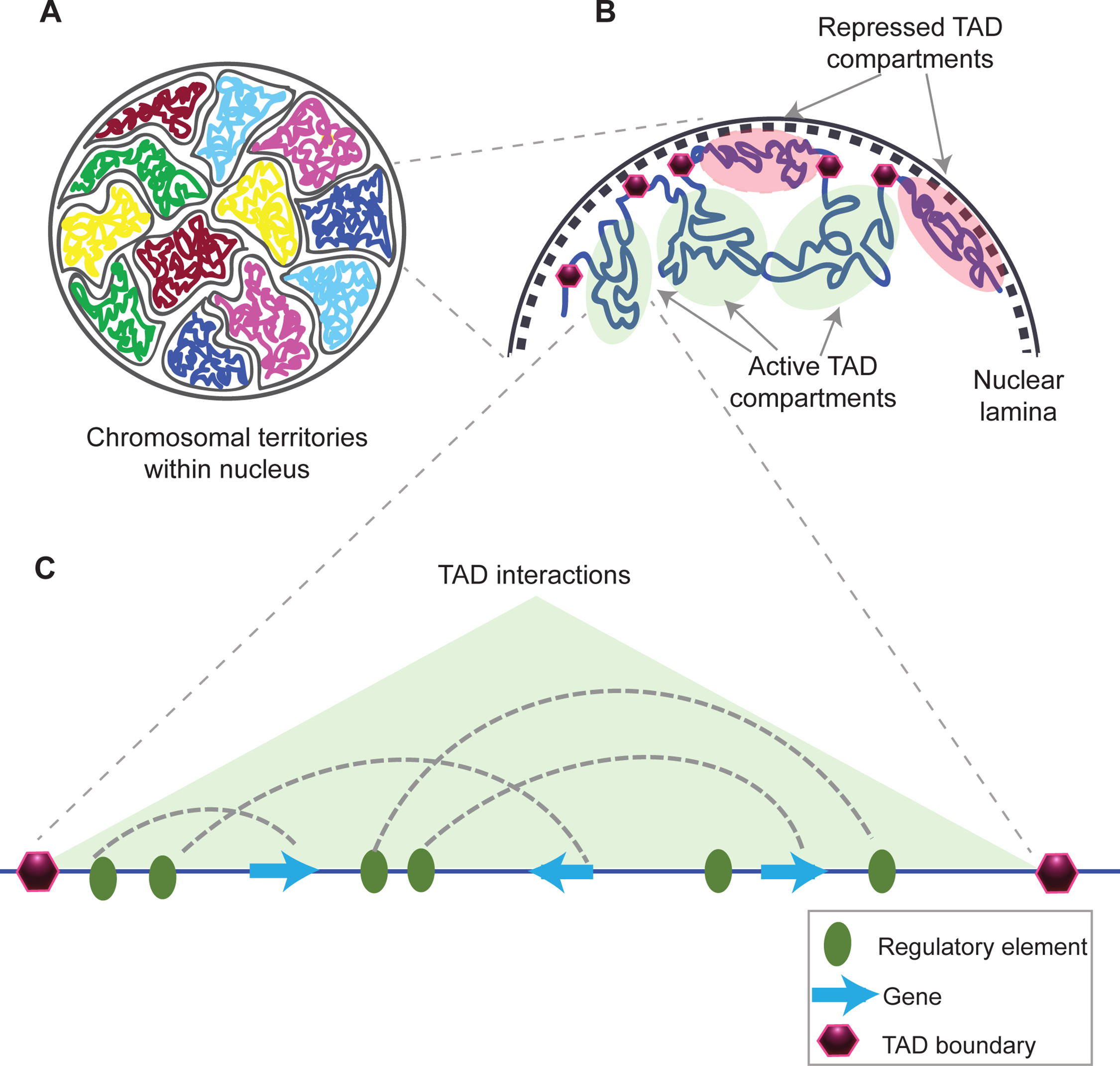
What are TADs?
-Highly conserved chromatin domains that shape functional chromosomal organization.
-Function not fully understood, their disruption lead to diseases
-
What are Chromosome territories?
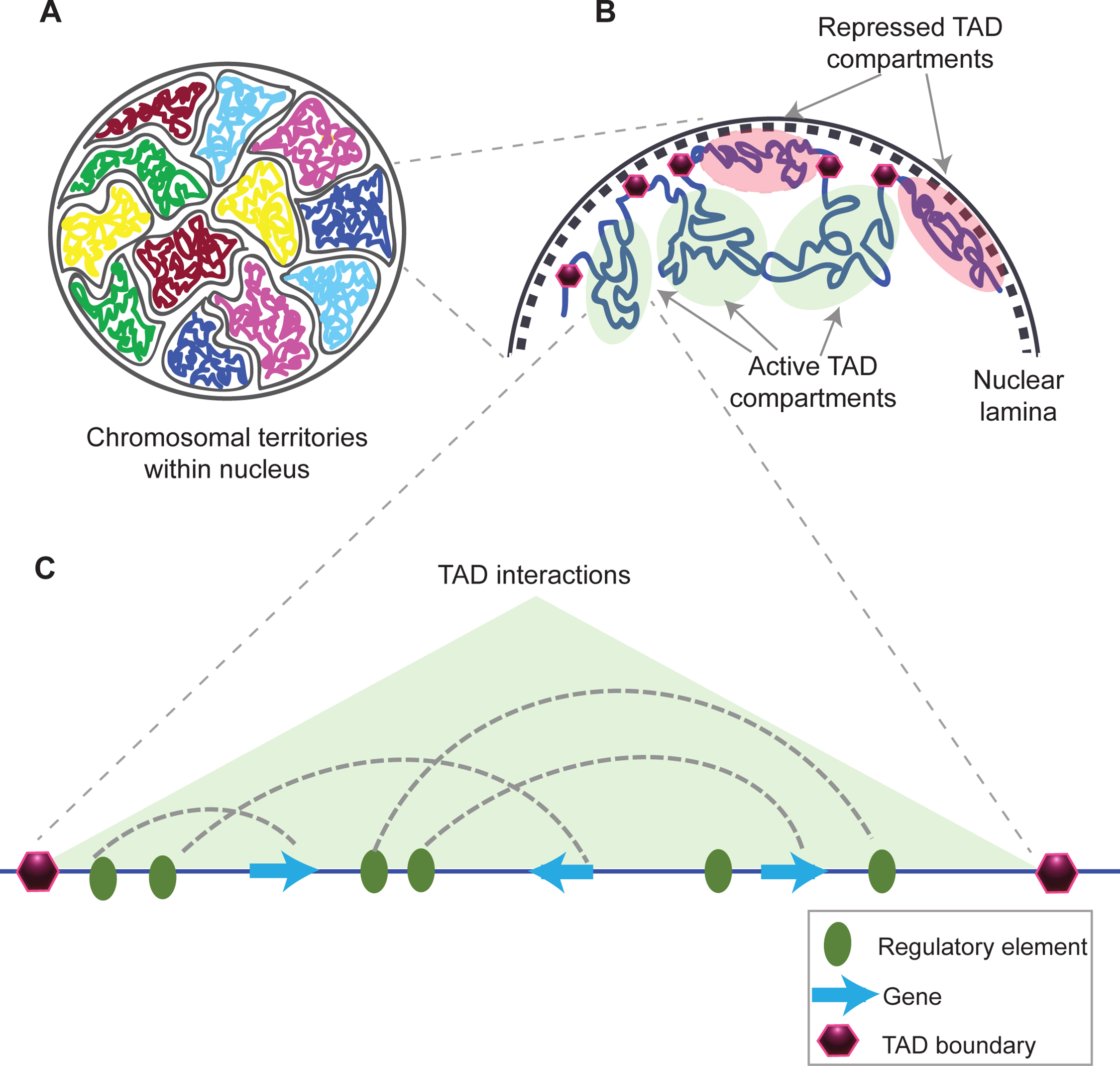
-Chromosome territories: non-overlapping domains/regions of the nucleus occupied by uncondensed chromosomes. They are composed of TADs.
-
What is Epigenetics?
- Heritable and reversible changes in gene expression which do not involve a change in the DNA sequence.
- Results from external or environmental factors or, as part of development program
-
What are the four different possible histone modifications that can occur? (mapu)
- Histone methylation
- Histone acetylation
- Histone phosphorylation
- Histone ubiquitination
-
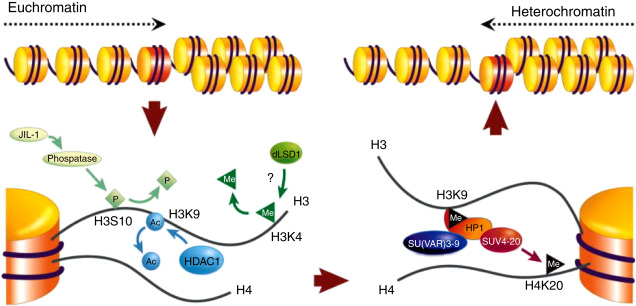
What is Histone Methylation? What about lysine residues?
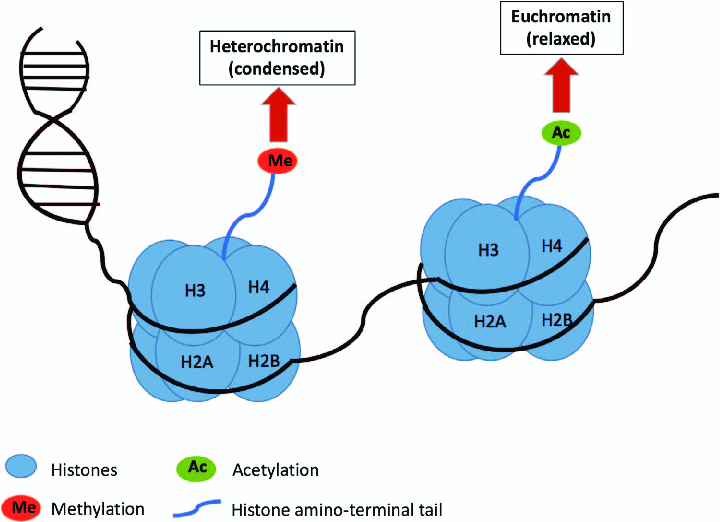
- Histone tails are methylated by histone methyl transferases (HMTs) and demethylated by histone demethylases (HDMs).
- Most common: mono-, di- or tri- methylation of lysines and arginines.
- Methylation of some lysine residues causes chromatin condensation (e.g. H3K9me3).
- Methylation of other lysine residues causes chromatin decondensation (e.g. H3K9me1).
- Effect may also vary if residue is mono-, di- or tri- methylated.
-
What is Histone Acetylation?
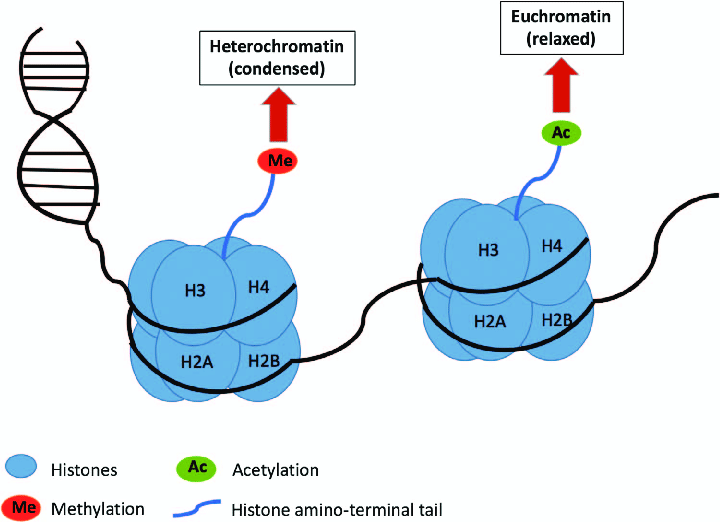
- Histone tails are acetylated by histone acetyl transferases (HATs) and deacetylated by histone deacetylases (HDACs).
- Most common: acetylation of lysines.
- Acetylated histones are generally associated with relaxed chromatin and gene expression and deacetylated histones with closed chromatin, silencing of genes.
- Aberrant acetylation is associated with several solid tumours and haematological malignancies.
- HDACi as emerging drugs in cancer treatment (e.g.vorinostat (SAHA) in T-cell lymphoma)
-
What is Histone Phosphorylation?
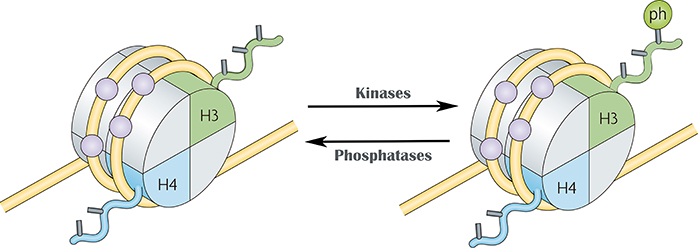
- Histone tails are phosphorylated by protein kinases and dephosphorylated by proteases.
- Can occur on serines, threonines and tyrosines.
- Phosphorylation of H3S10 and H3S28 is involved in chromatin condensation during mitosis and meiosis, as well as in chromatin relaxation linked to transcription activation.
-
What is HIstone ubiquitination?
-Addition of ubiquitin molecules to histone proteins
- Involves Ubiquitin ligases and deubiquitinating enzymes.
- Primarily on lysines of histones H2A and H2B of the histone tails
- H2Aub is more frequently correlated with gene silencing, while H2Bub is mostly associated with transcription activation.
-
What is the Histone code?
Combinations of post-translational modifications on the same histone’s tail/s.
-
What are code readers?
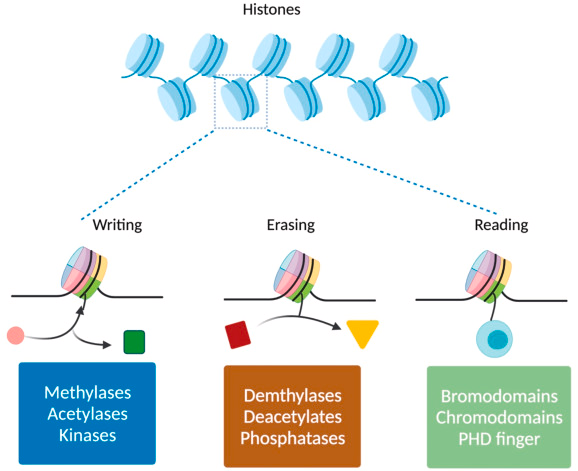
-Protein complexes that read combinations of marks (chemical mods):
-Homeodomains specifically recognize methylated residues, while
-Bromodomains bind acetylated residues.
-This read leads to chromatin remodelling (open or close)

