-
Draw a picture of and explain the components of a replication fork.
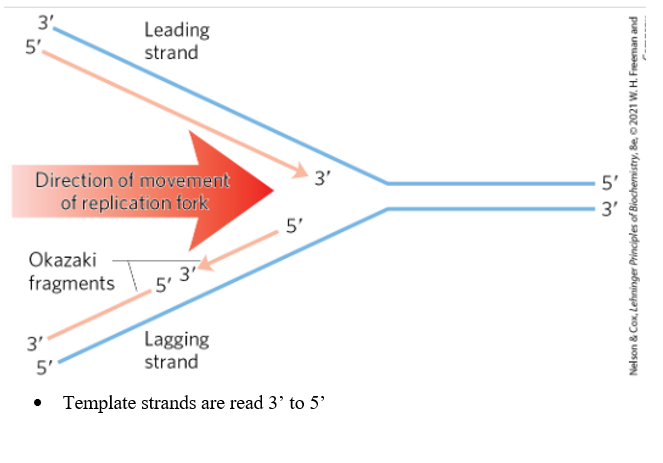
Replication forks are dynamic points where parental DNA is unwound and separated strands are replicated. This allows proteins involved in replication/synthesis to be able to access the template strands and allows both parent DNA strands to be replicated simultaneously (cuts time in half). Both ends of the bacterial chromosomes have active replication forks. Origin is where the loop opens, generating the fork. New strands are always synthesized 5' to 3'. Leading strand= continuously synthesizes 5' to 3' in the same direction that the replication fork. Lagging strand also synthesizes 5 to 3 but proceeds in the opposite direction of fork movement by Okazaki fragments. The template is read 3 to 5
-
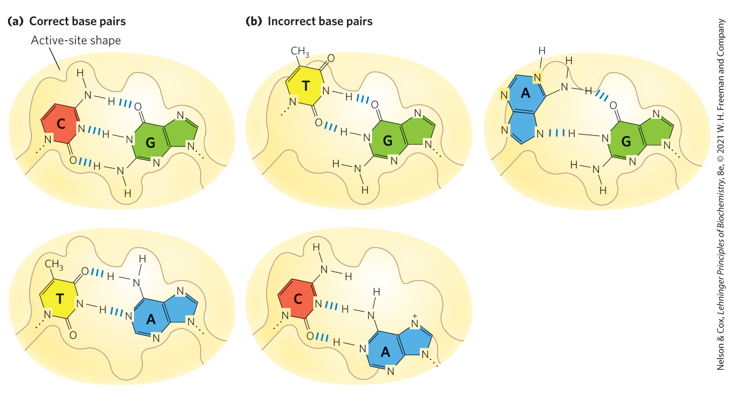
Be able to draw a G-C and A-T base pair.
-
Understand the reaction performed by DNA polymerase.
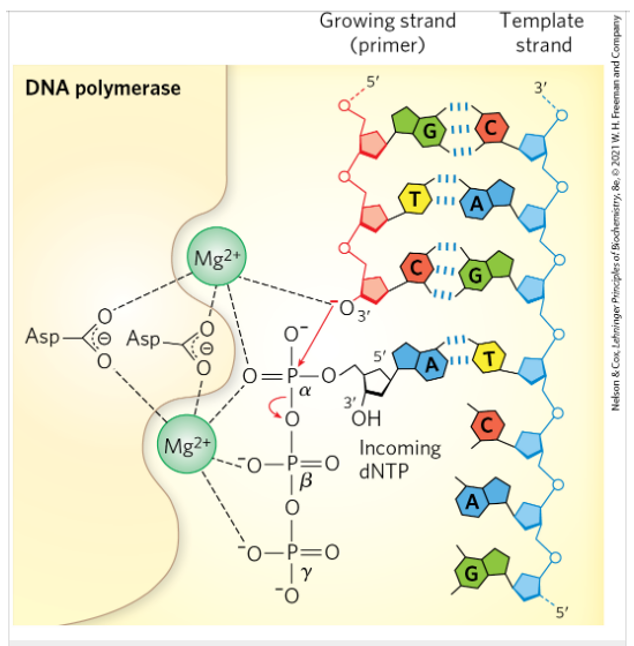
The catalytic mechanism for addition of a new nucleotide involved two Mg2+ ions coordinated to the phosphate groups of the incoming nucleotide triphosphate, the hyhroxyl group acts as a nucleophile, and 3 Asp residues. The Mg+ at the top facilitates the attack of the hydroxyl group of the primer on the phosphate of the nucleotide triphosphate; the other Mg2+ ion facilitates displacement of the pyrophosphate, both ions stabilizing the structure of the pentacovalent transition state. RNA polymerase uses a similar mechanism.
-
Understand the difference in roles between DNA polymerase I and DNA polymerase III.
DNA polymerase I: main enzyme for repair, removal of primers, and filling in the gaps in the lagging strand. Primary function is cleanup during replication, recombination, and repair (including nick translation, removing RNA primers, and connecting Okazaki fragments). 1 subunit. Has both 3 to 5 proofreading and 5 to 3 proofreading. 3-200 nucleotides added before polymerase dissociates.
DNA polymerase III: main enzyme catalyzing the 5 to 3 polymerization of DNA strands during replication. 9 subunits, only has 3 to 5 proofreading. 500,000 nucleotides added before dissociation.
-
Be able to describe the process of initiation of replication.
1. replication origin (oriC) serves as a binding site for key proteins
2. DnaA and DNA unwinding element (DUE) attach to these sites as well as three additional DnaA binding sites (I sites). There are also binding sites for proteins IHF, FIS, and HU
3. 8 DnaA proteins in ATP bound active form assemble to form a helical complex around the R and I sites, introducing a positive supercoiling effect
4. Strain on DNA strand leads to strand separation in the DUE
5. DnaC protein leads DnaB proteins onto the DNA seperated
-
Have a good knowledge of the composition of the replisome and how the various components coordinate to perform both leading strand and lagging strand synthesis.
1. helicases. Separate DNA into 2 strands that each act as a template. Require ATP, ie. DnaB
2. topoisomerases. Relive topological stress but on the DNA by strand separation. Also require ATP, ie. DnaB
3. DNA binding proteins. Stabilize separated strands, ie. SSB
4. Primases. enzymes that synthesize short segments of RNA to be present on the template. DnaG
5. DNA polymerase 1
6. DNA polymerase 3
7. RNase H1. Specialized nuclease, degrades RNA in RNA/DNA hybrids (removes RNA primers)
8. DNA ligases. seal nicks in DNA backbone from broken phosphodiester bonds after the RNA primers are removed by polymerase 1.
-
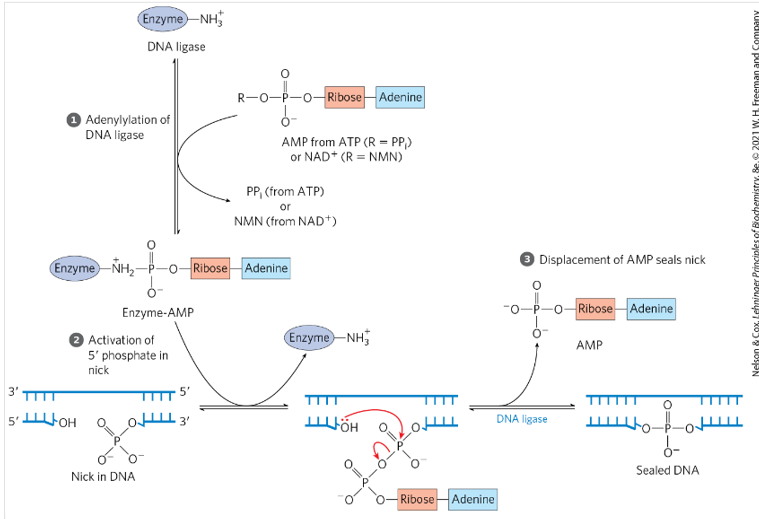
Know the chemistry involved in DNA ligation as shown in figure.
-
Be able to briefly describe termination of replication.
The two replication forks of the chromosome meet at a terminus region called Ter, which acts as a binding site for the protein Tus, which can arrest a replication fork from only one direction, preventing over replication by one fork in the event the other is delayed or halted. DNA circles are linked as catenanes, which must be separated by topoisomerase IV, separating them into 2 daughter cell chromosomes.
-
Describe how the various DNA repair systems in E. Coli as shown in Table 25.5. This includes knowing how DNA is damaged and the process used to fix those issues.
1. Mismatched repair. Corrected to reflect the template strand. Dam methylase methylates the repair site where incorrect nucleotides were placed. Immediatley after passage of replication, template strand is methylated while the new strand is not, permitting the two strands from being distinguished from one another.
2. Abnormal bases, alkylated bases, pyrimidine dimers. Base excision repair, removal of one base by DNA glycosylases that recognize common DNA lesions and remove them. AP endonucleases cut the DNA strand containing the AP site, DNA polymerase I replaces the DNA and DNA seals the nick.
3. Lesions causing structural strain. Nucleotide-excision repair, exonuclease hydrolyzes 2 phosphodiester bonds, one on either side of the distortion.
4. Pyrimidine dimers, methylation. direct repair, pyrimidine dimers caused by UV-induced reaction.
-
Choose 2 repair pathways and be able to describe them in detail.
1. Base excision repair. Uses DNA Glycosylases to recognize common DNA lesions and remove the affected base by cleaving the N-glycosyl bond, creating apurinic or apyrimidic site in the DNA called the AP site. AP endonuclease then cuts the DNA strand containing the AP site, the segment is removed. DNA polymerase I replaces the DNA, and DNA ligase seals the remaining nick.
2. Nucleotide-Excision Repair. Uses Excinuclease to hydrolyze 2 phosphodiester bonds, one on either side of the distortion caused by the lesion. Excised Oligonucleotides are released from the duplex and the resulting gap is filled by DNA polymerase I. DNA ligase seals the nick.
-
Know homologous genetic recombination including the names and functions of relevant proteins and how the mechanism takes place.
Involved the genetic exchanges between any 2 DNA molecules (or segments of the same molecule) that share an extended region or nearly identical sequence.
In bacteria can be used as a repair process when the replication fork collapses from encountering damaged site. 5' end is degraded, 3' is bound by a recombinase that pairs with a complementary sequence in the DNA complex. Branch migration moves the DNA branch and creates a Holliday intermediate (X-crossover). Nucleases cleave the holliday intermediate, and ligation restores the replication fork.
In Eukaryotes, this is required for proper chromosome segregation during meiosis, contributes to repair, and enhances genetic diversity in the population (crossing over) haploids.
-
yes
yes
-
Initation part 2
strands. This forms a tight complex with the ring-shaped, hexameric DnaB helicase, opening the DnaB ring
6. Two ring-shaped DnaB hexamers are loaded onto the DUE, one on each strand. DnaC ATP is hydrolyzed and dissociates the molecule.
7. DnaB migrates along the single strand of DNA in the 5' to 3' direction, unwinding the DNA as it travels, creating two potential replication forks.
8. DNA polymerase III is linked, and replication begins
9. Many molecules of single-stranded DNA binding proteins (SSB) bind to and stabilize the separated strands and DNA gyrase relieves the topological stress induced ahead of the fork by the unwinding reaction

