-
What are the markers for mature RNA and where do those RNA go?
Cap binding complex, Exon Junction Complex and poly-A-binding proteins. They go to the cytosol
-
What are the Eukaryotic initiation factors (eIFs) and what are they for?
5'cap bound by eIF4E and poly-A binding protein bound by eIF4G. To ensure the quality of the mRNA.
-
Where does mRNA quality control take place?
Cytosol
-
What checks for proper splicing in mRNA?
Exon junction complexes
-
What is Nonsense-Mediated mRNA decay?
mRNA surveillance system that checks for STOP codons in the wrong place(improper splicing).
-
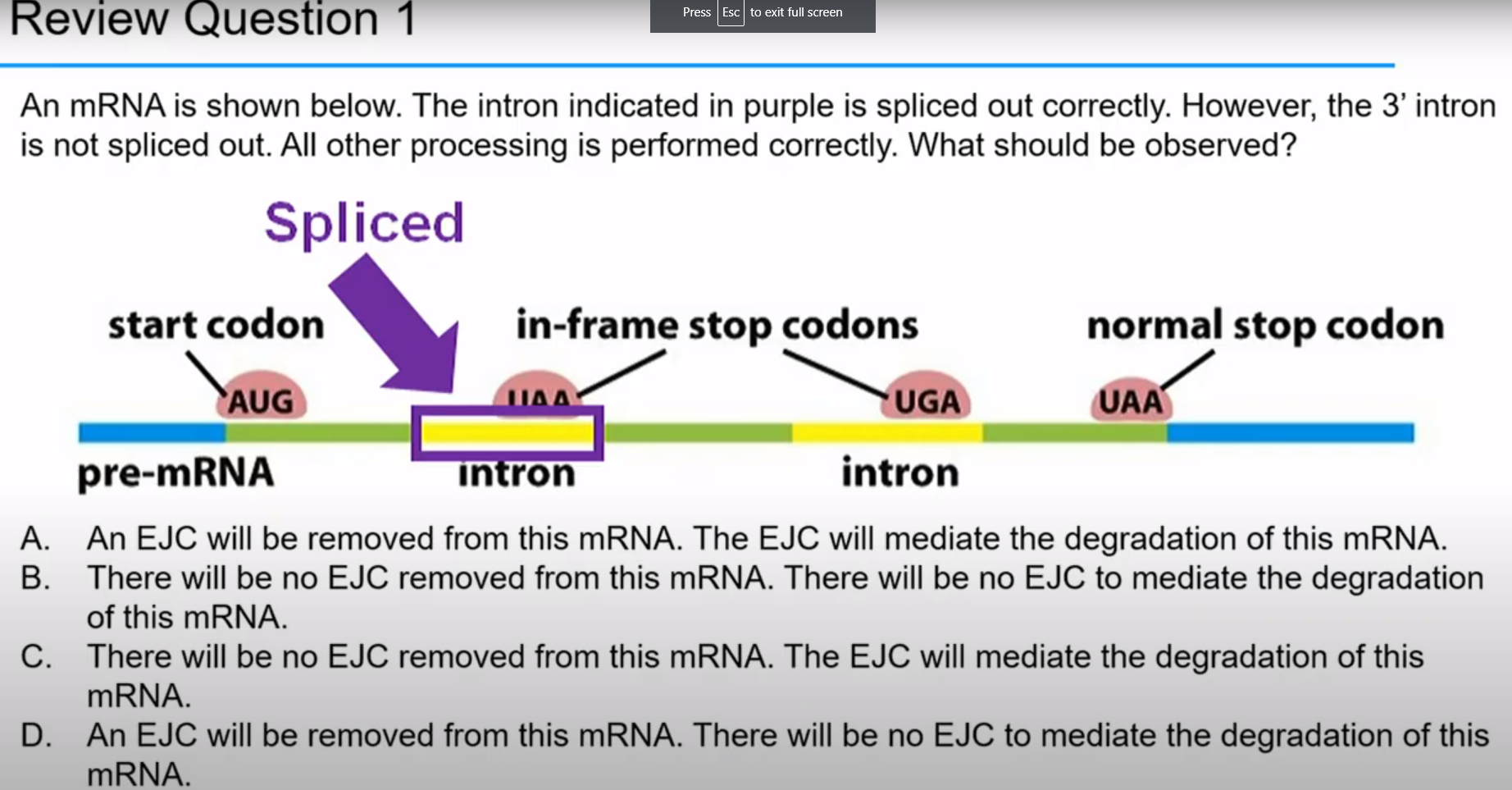
What happens when normal splicing occurs?
EJC's are displaced by moving ribosome and stop codon is in last exon. We have cap and poly-A-tail
-
What happens when abnormal splicing occurs?
EJC's are displaced by moving ribosome and stop codon is PREMATURE. EJC's remain on mRNA when ribosome reaches stop codon, mRNA is degraded by Upf proteins
-
D
-
What is used for quality control for broken mRNAs in prokaryotes?
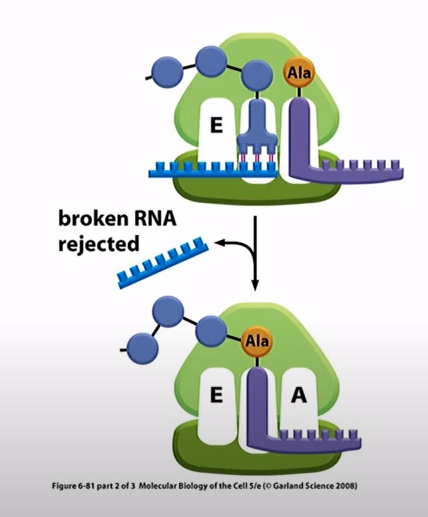
RNA called tmRNA that carries alanine amino acid and acts as both tRNA and mRNA. The ribosome would translate 10 codons from the tmRNA which acts as an mRNA. The 11 amino acid sequence will recognized by proteases that degrade the protein
-
What is the difference between mRNA stability in Prokaryotes and Eukaryotes?
In prokaryotes there are exonucleases that rapidly decay most mRNAs, while in Eukaryotes, mRNAs are more stable and degradation is regulated
-
How does mRNA stability work in Eukaryotes?
The poly-A-tail gradually shortens by an exonuclease(deadenylase) when mRNA reaches cytoplasm, it acts as a timer of mRNA lifespan
-
What happens when a poly-A-tail reaches critical length?
The cap gets bitten off or it continues to degrade and coding portion is eaten
-
What is the relationship between mRNA stability and Transferrin Receptors?
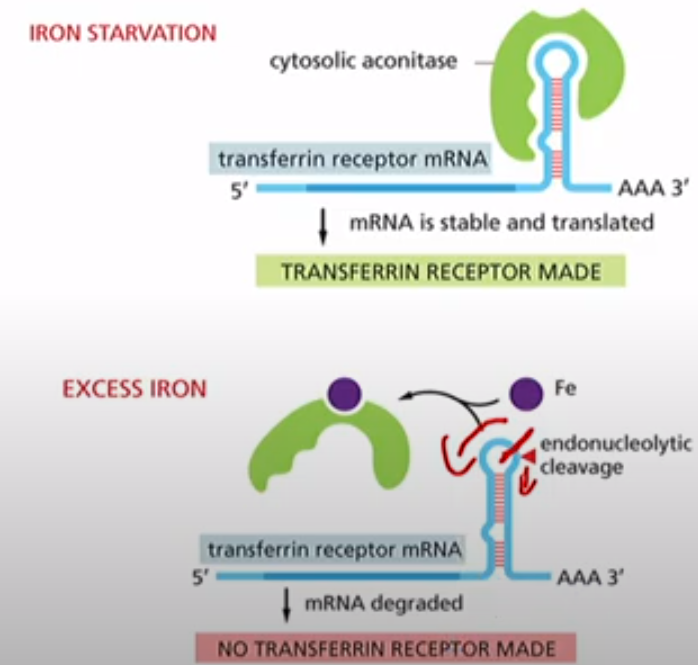
Transferrin Receptors import iron into the cell. Therefore during iron starvation you need to make more of them. So cytosolic aconitase binds to the transferrin receptor mRNA and keeps it stable so it can translate. When there is excess iron, it binds to cytosolic aconitase and now it cant bind to Transferrin mRNA. IT exposes the 3'UTR site that is susceptible to cleavage
-
What is the relationship between mRNA stability and Deadenylase
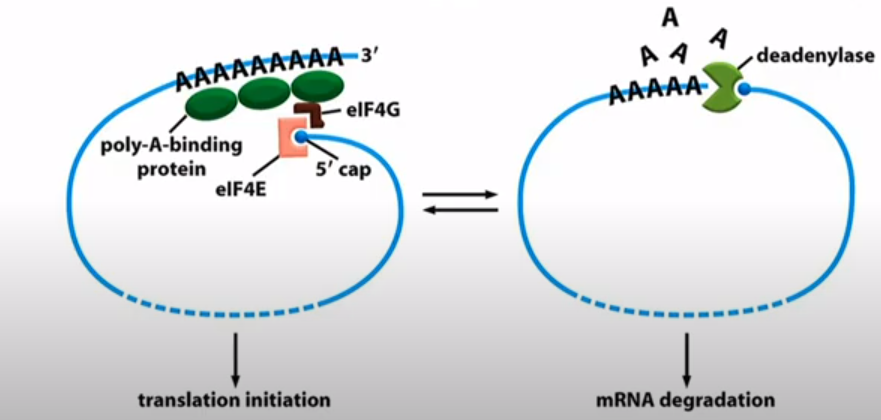
The initiation factors and the deadenylase compete for the 5' cap spot.
-
What are microRNAs(miRNAs)?
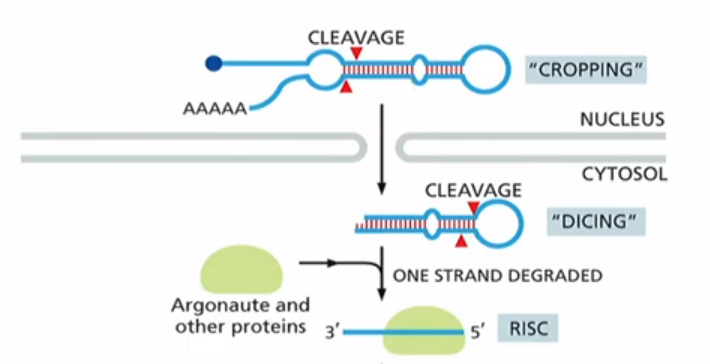
non-coding RNAs that regulate mRNA stability, they base pair with mRNAS. Are made by RNA polymerase ll and have 5' cap and poly-A tail. After processing, miRNA associates with protein complex called RNA-induced silencing complex (RISC)
-
What are two outcomes for the RISC complex
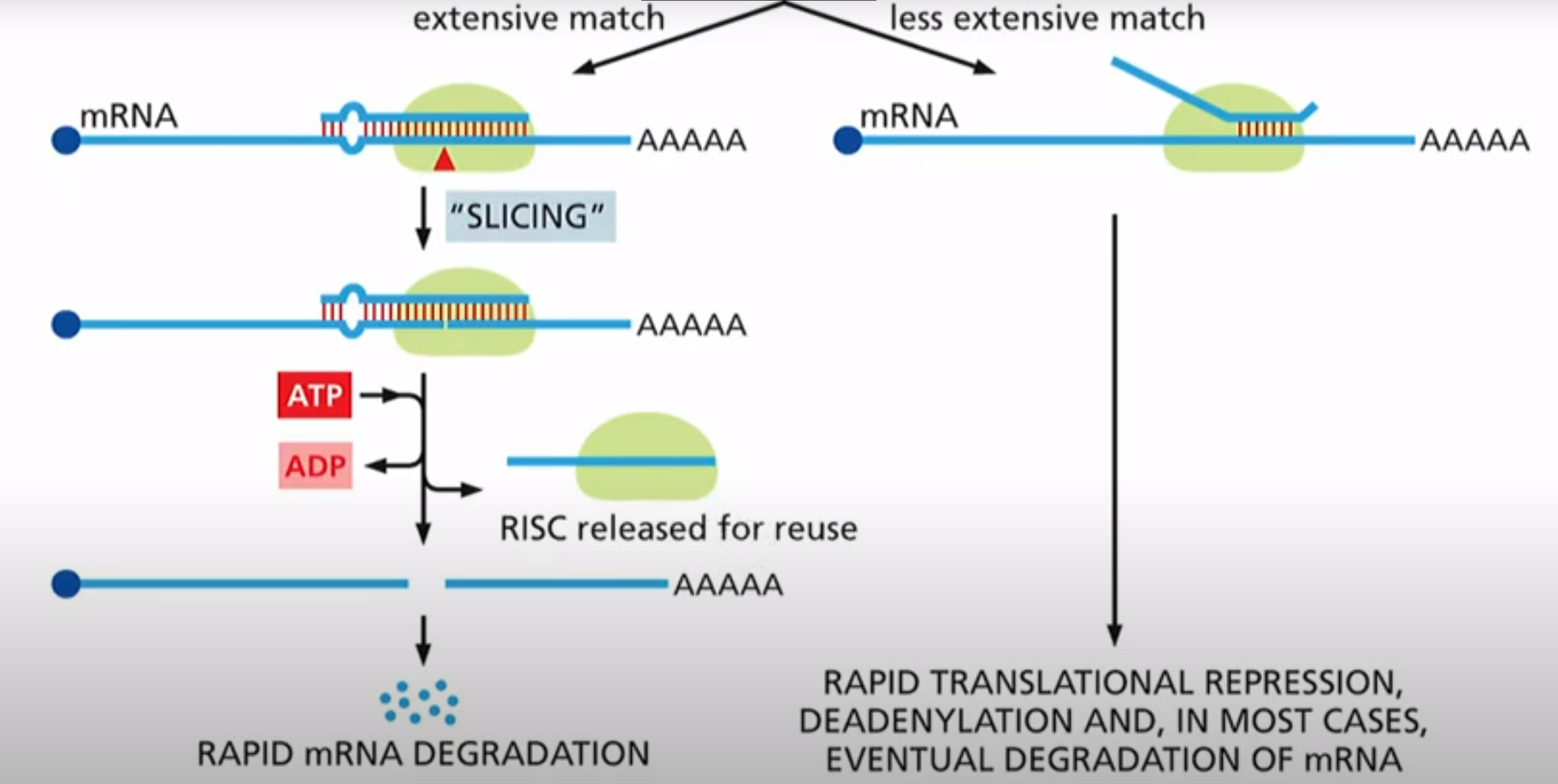
1. finds an mRNA it can bind to anti-parallel and complementary. It mediates slicing so it cuts that mRNA making it degrade. RISC is then reused
2. Blocks ribosome and mRNA cant translate. Eventually it degrades. RISC gets reused
This is called RNA interference (RNAi)
-
How does RNAi and siRNAs interact?

Virus make double stranded RNAs. RNAi destroys double stranded RNA through Dicers. The leftover parts are siRNAs. Make a RISC out of those and inhibit viral particles
-
How does CRISPR-Cas immunity work?