What is a microarray? (1)

An ordered assembly of nucleic acids immobilized on a solid support, often glass (like a microscope slide).
What are microarray probes made of? (2)
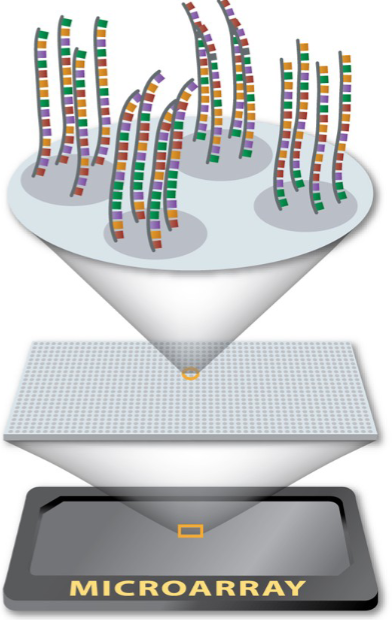
Probes are short pieces of single-stranded DNA.
They are oligonucleotides immobilized on the surface of the array.
What does each spot on a microarray consist of? (1)
Thousands of probes with the same sequence.
What are the primary applications of microarrays? (3)
Gene expression (transcriptomics).
SNP genotyping (SNP arrays).
Structural variant detection (array CGH).
What is replacing many microarray applications? (2)
Next Generation Sequencing (NGS) protocols like RNA-Seq, WES (Whole Exome Sequencing), and WGS (Whole Genome Sequencing).
Are Next Generation Sequencing protocols widely used in clinical settings yet? (1)
No, not widely used in the clinic yet.
What are the two types of arrays mentioned? (2)
Spotted arrays.
Printed arrays.
What are the examples of structural variant detection in microarrays? (1)
Array Comparative Genomic Hybridisation (aCGH). - Detects CNVs such as deletions or duplications by comparing the DNA of a test sample with a reference sample.
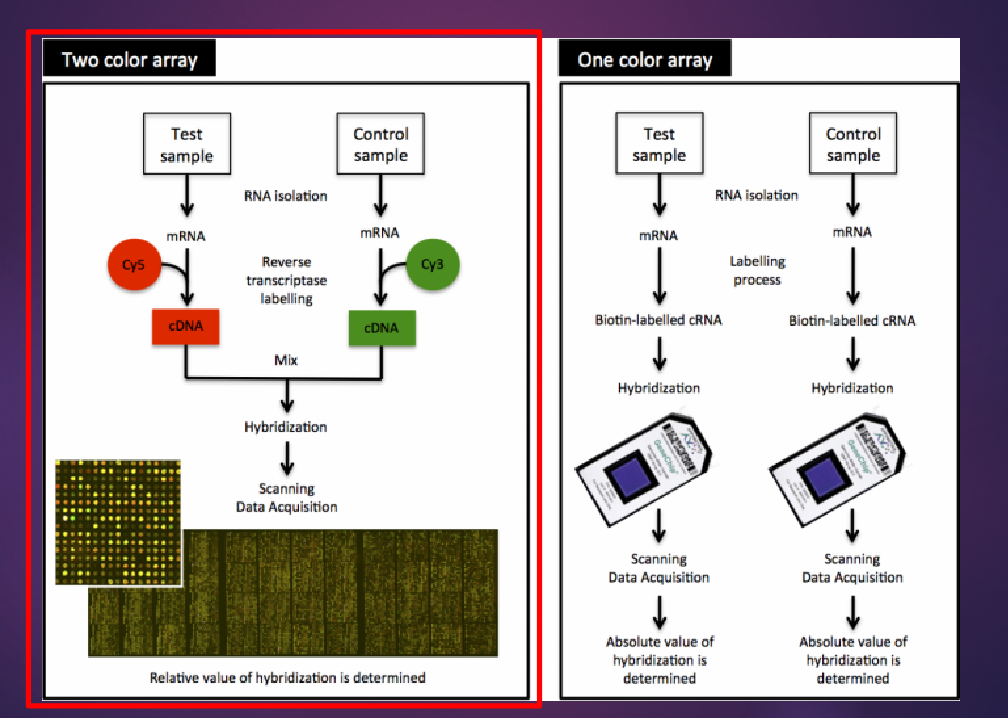
What is RT-PCR used for in microarray studies? (1)
Validation of results obtained from gene expression arrays.
What is the transcriptome? (1)
The complete set of RNA transcripts produced by the genome.
What is transcriptomics used for? (2)
To discover the expression levels of all genes in a sample.
To determine which genes are expressed at different levels between two sample types.
What can transcriptomics help you discover? (1)
The biology of your samples by analyzing gene expression patterns.
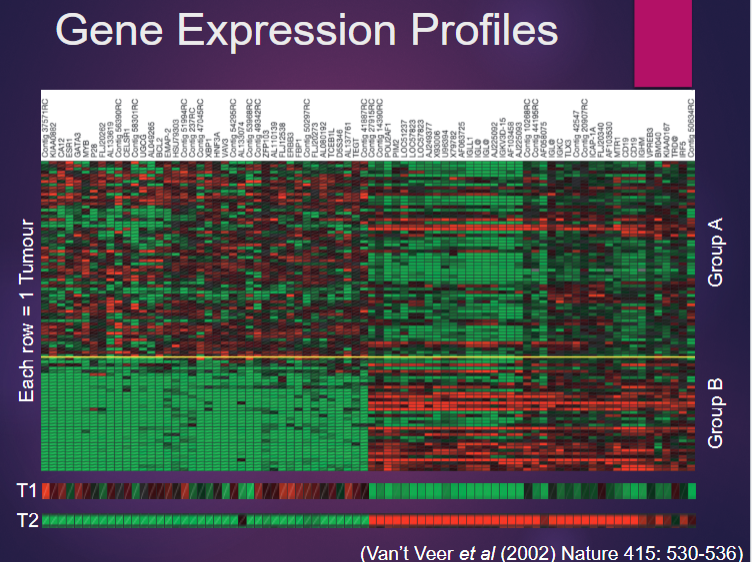
How can transcriptomics be used to classify samples? (1)
It can classify samples based on their gene expression profiles.
How can transcriptomics help predict sample class? (1)
It helps predict the class or type of a sample based on its gene expression levels.
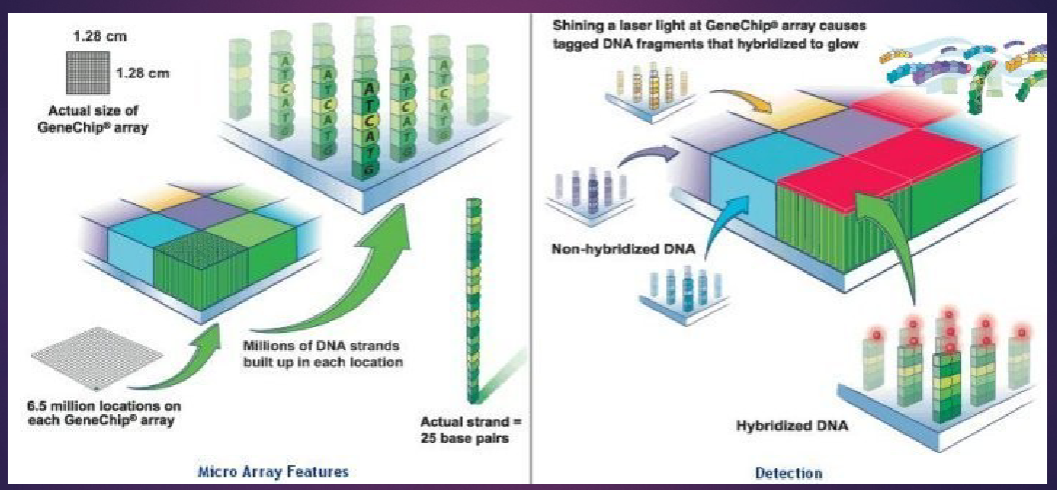
What do gene expression microarrays contain? (3)
Lots of copies of the same probe in a spot
Each spot gives the relative expression for one transcript
Detects all known transcripts in one sample
Picture demonstrating Expression Profiling Workflow:
Picture demonstrating data analysis workflow:

What is Expression Analysis and why is it important in genomics? (2)
Expression analysis involves measuring gene expression levels in samples.
It is important for understanding gene function and identifying disease mechanisms.
What is Normalisation in gene expression data, and what is its significance? (2)
Normalisation is the process of adjusting gene expression data to account for technical variations.
Its significance lies in ensuring comparability across samples for accurate analysis.
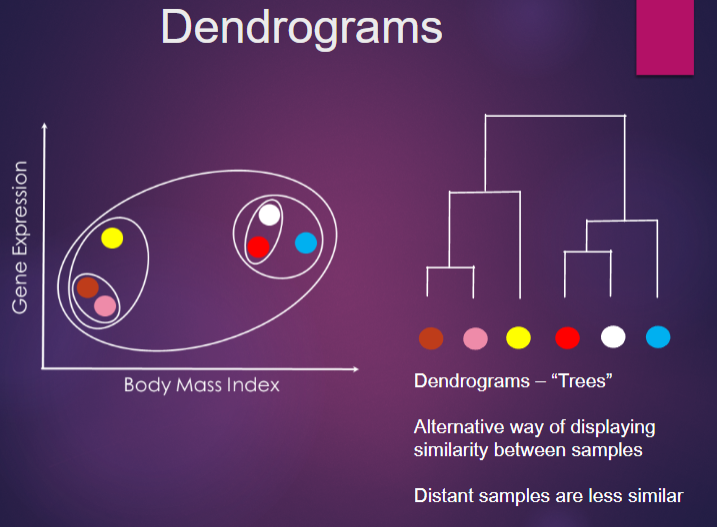
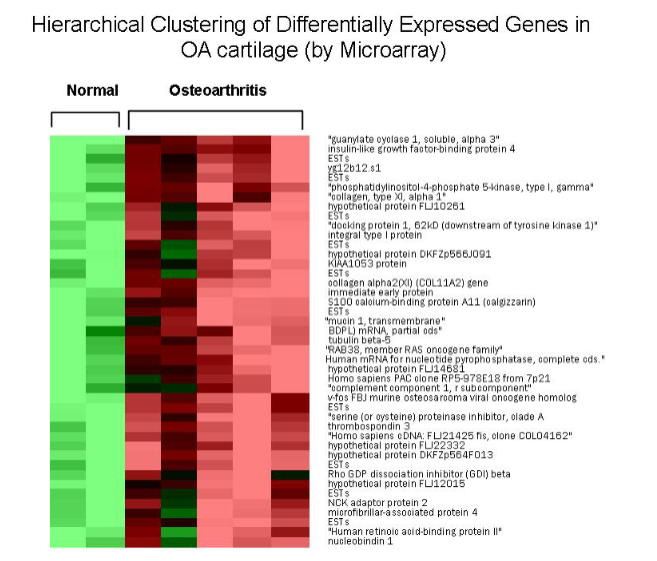
What is Hierarchical Clustering, and how is it used in genomics? (3)
Hierarchical clustering is a statistical method used to group genes or samples based on expression profiles.
It is used in genomics to identify patterns and relationships in gene expression data, aiding in the discovery of co-regulated genes.
This helps researchers make sense of complex datasets.
What is Gene Filtering, and why is it necessary in expression analysis? (2)
Gene filtering involves selecting a subset of genes based on specific criteria.
It is necessary to reduce data complexity and focus on the most relevant genes for further analysis.
What are Statistical Tests in the context of gene expression studies, and what is their purpose? (3)
Statistical tests determine the significance of observed differences in gene expression between groups.
Their purpose is to validate findings and ensure results are not due to random chance.
This is critical for drawing reliable conclusions from the data.
What does it mean to Generate a Gene List, and how is it utilized in research? (2)
Generating a gene list involves compiling genes that meet specific criteria from the analysis.
This list is utilized for downstream analyses, such as pathway enrichment and biological interpretation.
What is Biological Interpretation in the context of gene expression analysis, and why is it important? (2)
Biological interpretation involves linking gene expression data to biological processes and pathways.
It is important because it provides context to the data, helping researchers understand the biological implications and potential applications of their findings.
What is Clustering and what are its key characteristics? (2)
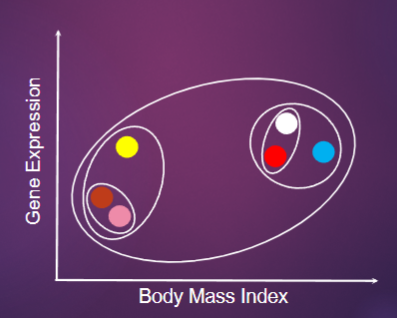
Clustering organizes data with similar patterns into classes.
Objects within a class are more similar to each other than to objects outside the class.
Picture demonstrating dendograms:
Picture demonstrating Hierarchical Clustering of Differentially Expressed Genes in OA cartilage (by Microarray):
Picture demonstrating gene expression profiles:
What are Data Repositories and how do they maximize the utility of microarray experiments? (3)
Data repositories maximize the utility of microarray experiments by allowing users to share data.
They enable researchers to use other people's data, enhancing collaborative research.
If users provide the Minimum Information About a Microarray Experiment (MIAME), it becomes easier to compare results.
Name two examples of data repositories. (2)
ArrayExpress - run by EBI (European Bioinformatics Institute)
GEO - run by (Gene Expression Omnibus), NCBI (National Center for Biotechnology Information, USA)
What is Quantitative-PCR (qPCR) and how is it used to confirm microarray results? (3)
qPCR, also known as RT-PCR, is a technique used to confirm microarray results by quantifying gene expression levels.
It involves amplifying the DNA of a gene of interest and comparing it to a housekeeping gene for normalization.
The assessment can be done at the transcript level, measuring DNA, RNA, and protein levels through the use of reverse transcriptase.
How can RT-PCR be made quantitative? (2)
RT-PCR can be made quantitative by counting the number of copies of amplified DNA present.
This counting is achieved by using fluorescent molecules, often referred to as “tags,” which allow for the detection of the amplified DNA.
Picture demonstrating Exponential Amplification in PCR:
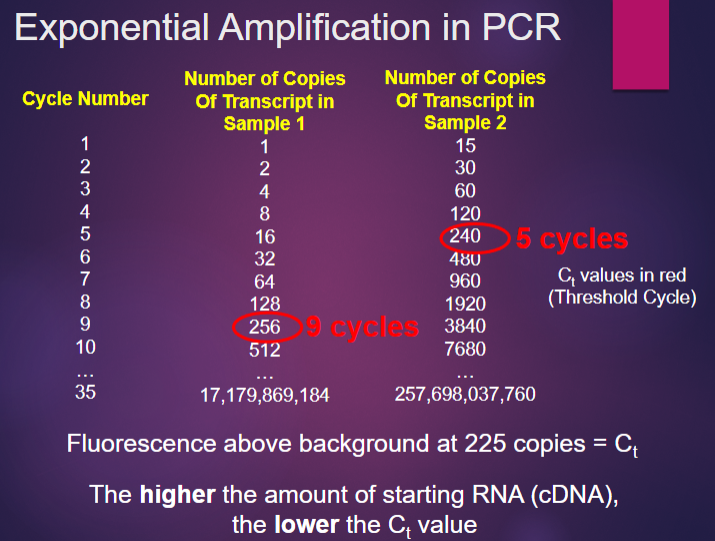
How do you count the number of amplified molecules present in PCR? (2)
Include a dye in the PCR reaction mix that fluoresces when it binds to double-stranded DNA, such as an intercalating dye like SYBR Green.
Alternatively, label a probe in the PCR that only fluoresces when it is incorporated into the PCR product, such as TaqMan.
Picture demonstrating DNA amplification Plot for qPCR:
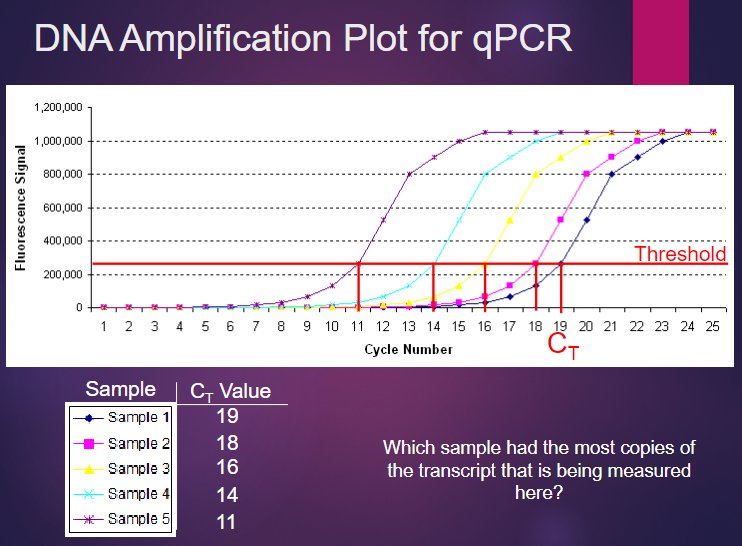
Why is qPCR used in gene expression analysis? (3)
qPCR is used to independently confirm differences in RNA levels between samples.
Probe binding can be noisy, and differences detected may not be real, especially for small differences (less than 2-fold).
While RNA-Seq provides a more accurate measure of RNA transcript abundance and is more reproducible over a wider range of concentrations, it is generally more expensive.
Picture demonstrating Predicting Breast Cancer Recurrence:
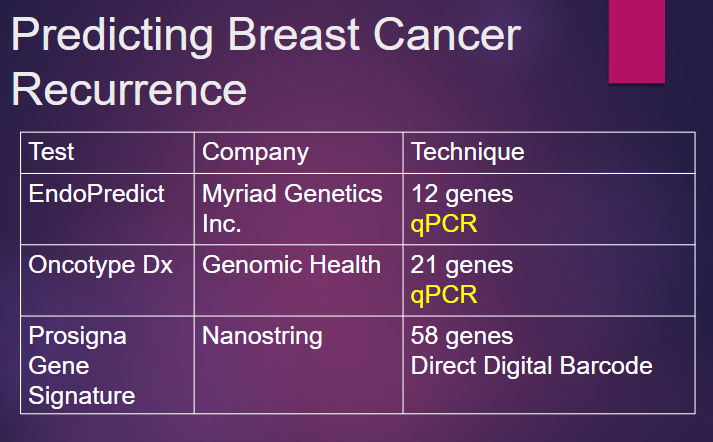
What is EndoPredict Risk Estimation in the context of breast cancer, and how does it guide treatment decisions? (3)
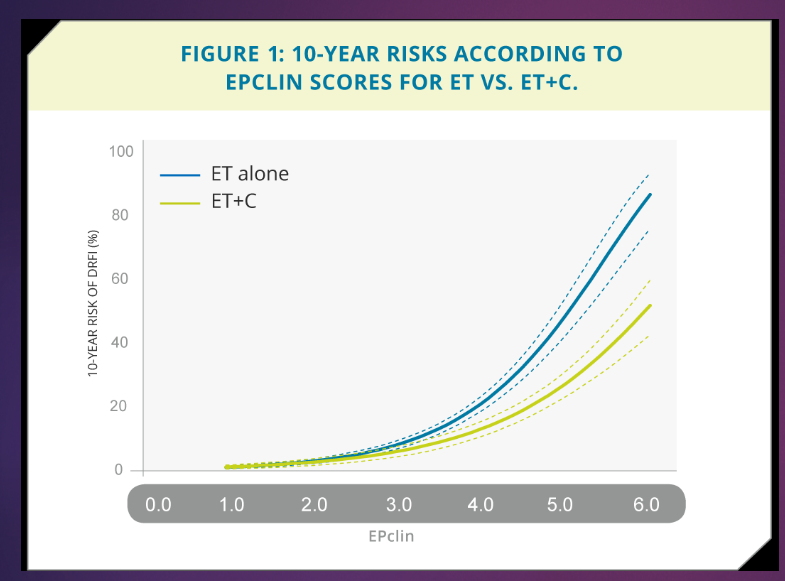
At a low score (EPclin < 3), endocrine therapy (ET) alone is sufficient for treatment.
At higher scores, combination therapy (ET+C) is clearly beneficial compared to ET alone.
This test ensures that only patients who will benefit from chemotherapy receive it, optimizing treatment plans.
What does DFRI stand for? (1)
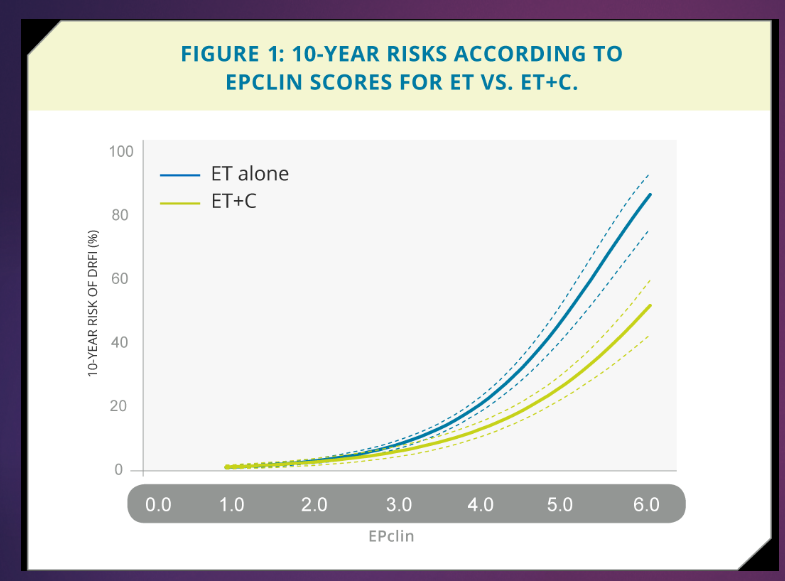
DFRI stands for distant recurrence-free interval.
What are Single Nucleotide Polymorphism (SNP) microarrays and how are they used in Genome-wide Association Studies (GWAS)? (3)

SNP microarrays enable Genome-wide Association Studies by allowing the genotyping of large numbers of SNPs in a large number of subjects.
This is achieved by using microarrays that hybridize with genomic DNA adjacent to SNPs rather than RNA transcripts.
The SNP is then extended by one base that is fluorescently labeled and detected using a high-definition scanner.
What is contained in a spot on a SNP microarray? (2)
A spot contains many copies of the same single-stranded oligonucleotide, referred to as a “probe.”
Each probe is designed for genotyping one specific SNP.

How do SNP microarrays work? (3)

SNP microarrays consist of a grid of spots, each containing probes specific to a particular SNP.
When genomic DNA is applied to the microarray, it hybridizes with the complementary probe at each spot.
The hybridized DNA is then extended by one fluorescently labeled base, allowing detection via a high-definition scanner, which identifies the presence or absence of specific SNPs based on the fluorescence signals.
What are genotyping microarrays and their key features? (3)
Each spot on a genotyping microarray provides the genotype for one specific SNP.
Up to 5 million spots can be present per sample on the array, allowing for extensive analysis.
This technology enables genome-wide analysis, facilitating large-scale genetic studies.
How are SNP genotypes determined from microarray data? (3)
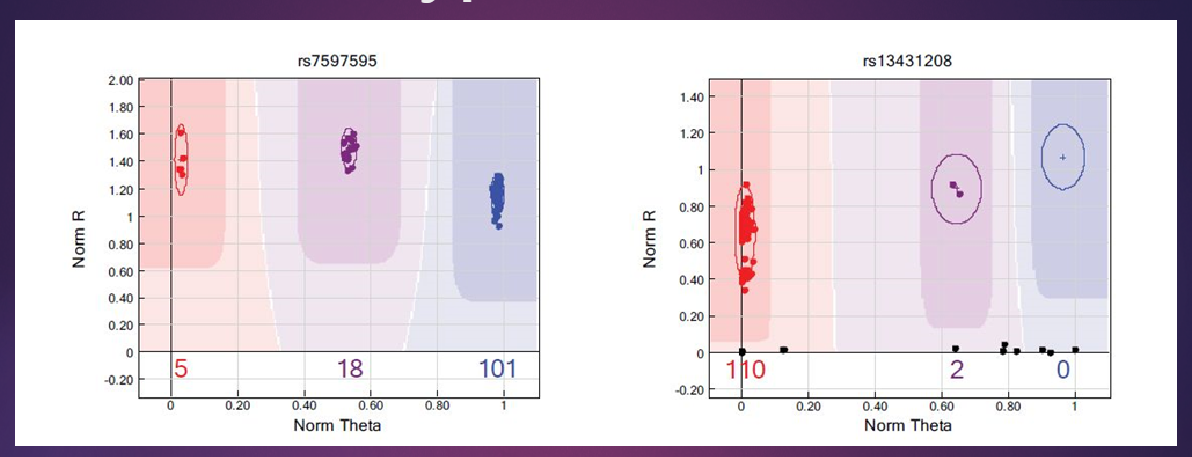
Probes on the microarray detect specific SNP variants by binding to them, with each variant having a different color signal.
The software analyzes the color intensity from each probe and assigns the corresponding genotype (e.g., homozygous reference, heterozygous, or homozygous alternative).
While most SNPs are processed automatically, some are manually checked to ensure accuracy.
Picture demonstrating Genome Wide Association Study:
What is Array-CGH and what is its purpose in genomic analysis? (3)

Array-CGH is a genomic hybridization technique used to detect copy number variations (CNVs) across the genome.
It involves comparing the genomic DNA of a test sample to a reference sample to identify deletions or duplications of DNA segments.
This method is crucial for understanding genetic disorders, cancer, and other diseases by providing insights into genomic alterations.
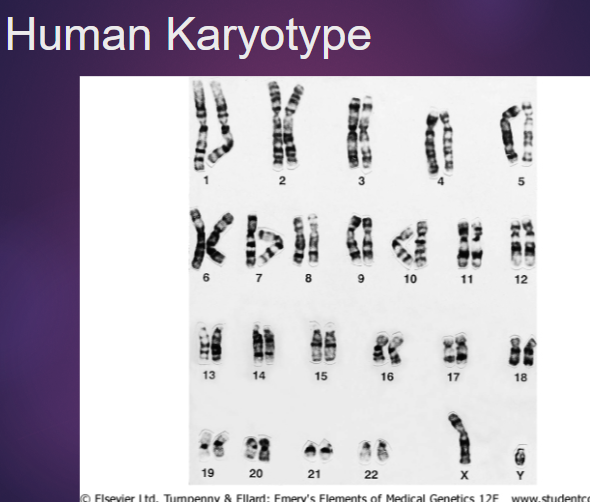
Picture demonstrating the human karyotype:
What are Structural Variants (SV) and Copy Number Variants (CNV), and how do they differ? (3)
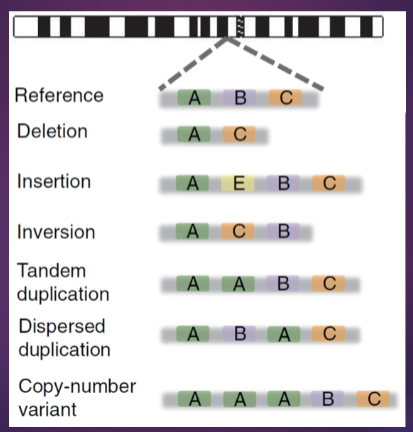
Structural variants (SV) refer to large alterations in the genome, such as inversions, translocations, or large deletions and duplications.
Copy number variants (CNV) are a specific type of structural variant that involves changes in the number of copies of a particular DNA segment, leading to either gains (duplications) or losses (deletions) of genetic material.
While all CNVs are considered structural variants, not all structural variants are CNVs; SVs encompass a broader range of genetic changes.
What is Array Comparative Genomic Hybridization (aCGH) and what are its key applications? (3)
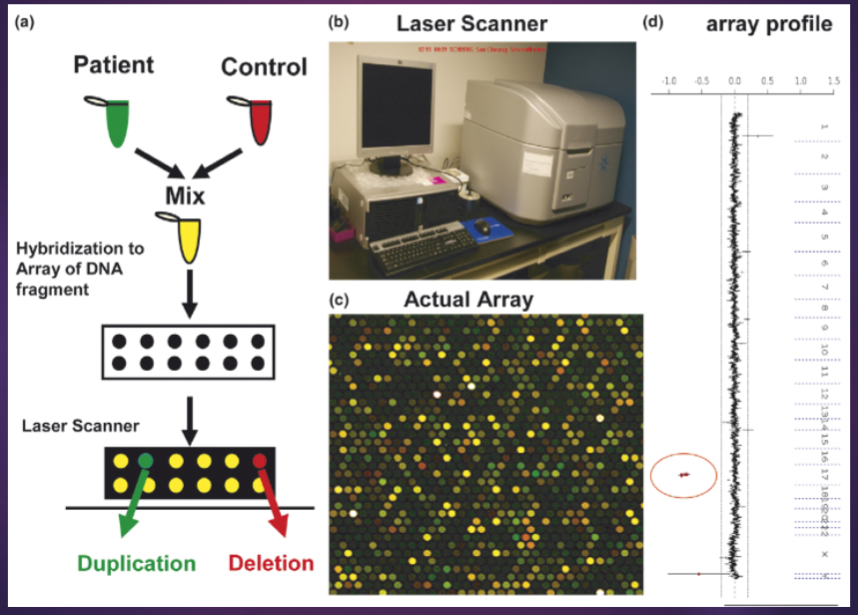
aCGH is a molecular cytogenetic technique used to detect and quantify genomic copy number variations (CNVs) across the genome.
It involves comparing labeled DNA from a test sample to a reference sample, allowing for the identification of deletions, duplications, and other structural variations.
Key applications of aCGH include cancer research, prenatal testing, and the study of genetic disorders, providing insights into genomic imbalances associated with various diseases.