What is the structure of DNA? (2)
A macromolecule consisting of a linear strand of nucleotides
Double-stranded formed by binding complementary strands
What is the DNA packing problem? (1)
The DNA packing problem refers to the challenge of fitting an extremely long strand of DNA into a small space, like the nucleus of a cell. Its a 'problem' in theory
What are the components of the exome? (2)
Genes
Gene structure
What processes are involved in gene expression? (2)
Transcription
Translation
What is meant by the 3D Genome? (1)
The spatial organization of DNA within the nucleus
Draw the structure of a nucleotide (3)
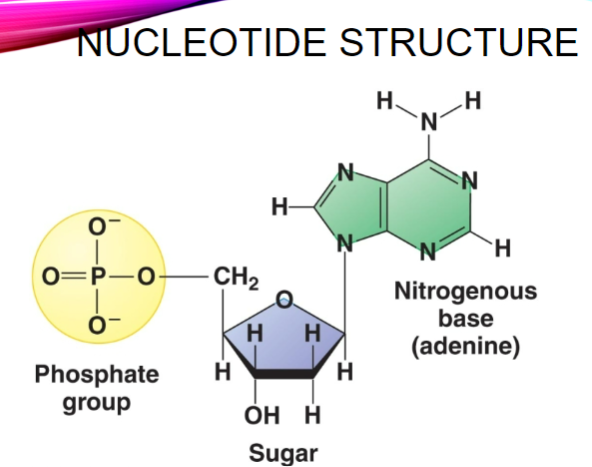
Picture demonstrating the structure of DNA
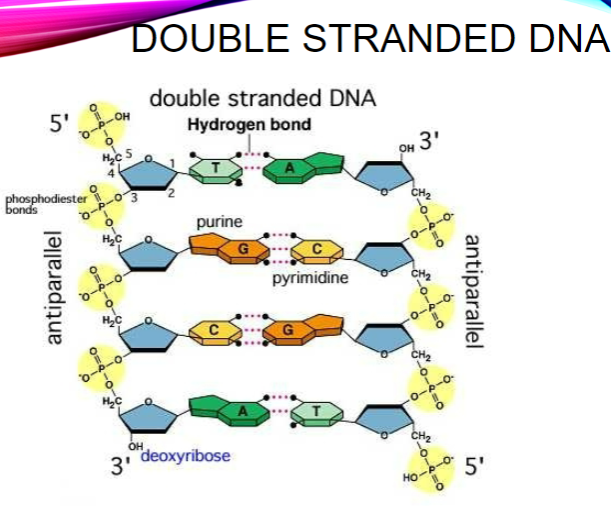
What indicates the directionality of single-stranded DNA? (1)
The 5’ and 3’ carbons
Where does the numbering of carbons in DNA start? (1)
At the carbon closest to the base
What is the sequence direction of DNA by convention? (1)
5’ -> 3’
What is the DNA sequence provided? (1)
TAA
What are the two orientations of DNA strands? (1)
Antiparallel
How are the bases arranged in the structure of DNA? (1)
Stacked
What are the two types of grooves present in DNA? (2)
Major
Minor
What is the size of the human genome in base pairs? (1)
3 x 10^9 base pairs (3 Gb)
How many genes are approximately contained in the human genome? (1)
~20,000 genes
What is the trend regarding the number of genes in simpler organisms? (1)
Simpler organisms tend to have fewer genes.
How many genes do flies, yeast, and bacteria have approximately? (2)
Flies: 10,000
Yeast: 4,000
Bacteria: 1,000
Is genome size strongly related to the complexity of an organism? (1)
No, genome size is not strongly related to complexity.
What are two examples of organisms with large genome sizes that are not necessarily complex? (2)

Marbled lungfish: 130 Gb
Paris japonica: 149 Gb
What is the approximate length of DNA in a single human cell? (1)
~2 meters of DNA
How many cells are there approximately in the human body? (1)
37.2 trillion cells
What is the total length of DNA in the human body in meters? (1)
7.44 x 10^13 meters of DNA
How does the total length of DNA compare to the diameter of the Solar System? (1)
It is twice the diameter of the Solar System.
What are histones? (1)
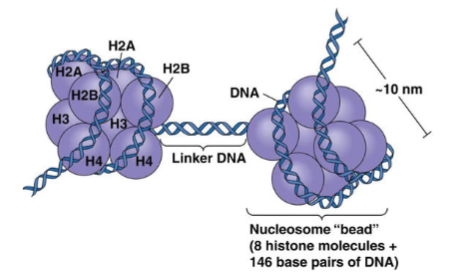
Positively charged (basic) proteins that bind DNA.
How many histones make up a nucleosome? (1)
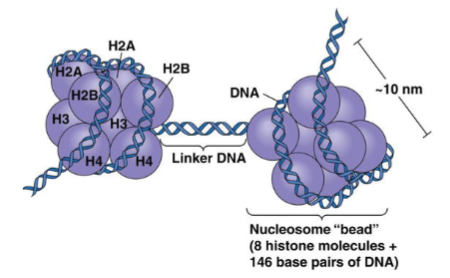
Eight histones: 2x(H2A + H2B + H3 + H4).
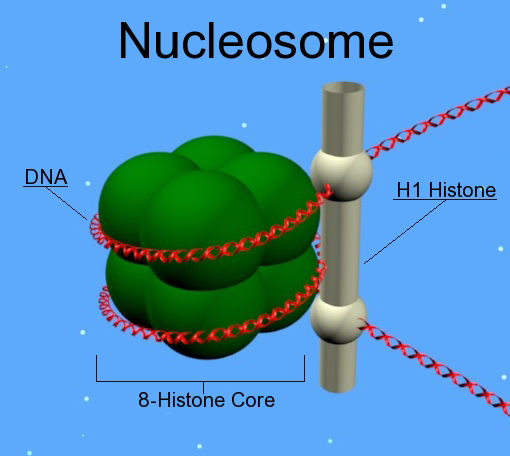
What is the role of histone 1? (1)
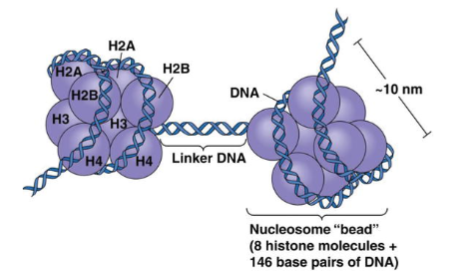
Histone 1 binds the linker DNA between nucleosomes.
Describe DNA packing (5 steps, 1 mark for each step)
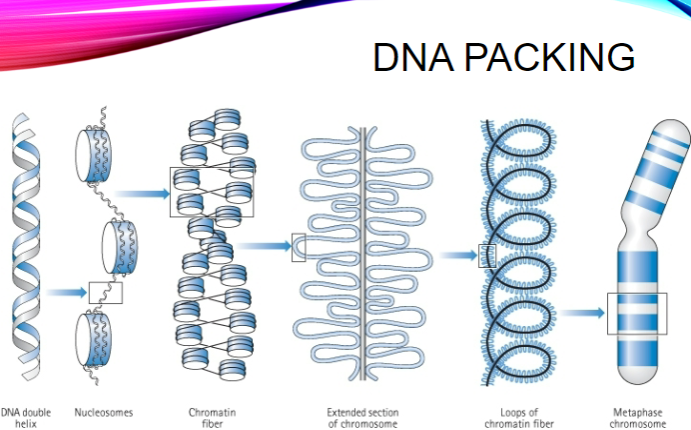
DNA forms a double helix.
The double helix coils into nucleosomes (DNA wrapped around histone proteins).
Nucleosomes pack into chromatin fibers.
Chromatin fibers fold into extended sections of a chromosome.
The extended sections form loops to create the condensed metaphase chromosome.
What is the exome? (1)
The sum of all gene sequences that code for a protein
What is the size of the coding sequences in the exome? (1)
Approximately 37 Mbp (1.2% of the genome).
What is the size of all gene sequences in the exome? (1)
Approximately 60 Mbp (2% of the genome).
How do we define a gene? (2)
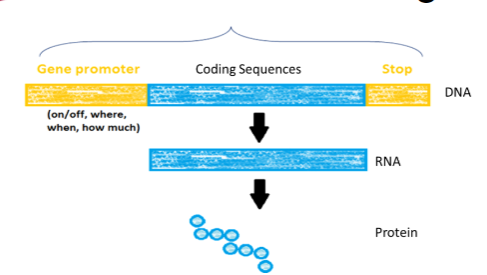
A gene is a segment of DNA that contains the instructions to produce a specific RNA molecule. This RNA molecule may function directly (e.g., rRNA, tRNA) or be translated into a protein.
A gene includes not only the coding sequences (exons) that specify the protein but also the non-coding sequences (introns) and regulatory elements (e.g., promoters, enhancers, silencers) that control when, where, and how much of the gene product is made.
What is the size of the globin gene? (1)
1.8 kb.
What is the size of the dystrophin gene? (1)
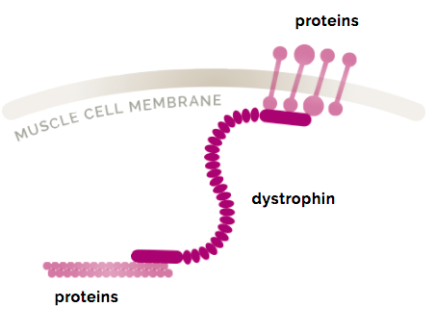
2.4 Mb.
What do intergenic regions contain? (5)
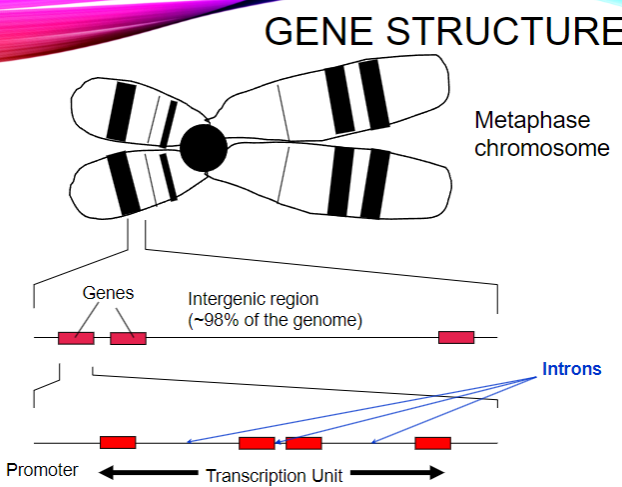
Regulatory elements.
Sequences of no known function, such as repetitive DNA, endogenous retroviruses, and pseudogenes.
What does it mean that genes often cluster in families? (2)
It allows for coordinated gene regulation.
It may reflect evolutionary history.
What is the range in number of introns in genes? (1)
From 0 to over 300.
What is the range in size of introns? (1)
From 30 bp to 1 Mbp.
Can introns contain other genes? (1)
Yes, some introns contain other genes.
Which type of genes typically lack introns? (1)
Genes that code for histone proteins.
What purpose do introns serve? (2)
They may play roles in gene regulation and alternative splicing.
They can provide a means for evolutionary changes and increased protein diversity.
What is the purpose of the regulatory element in the promoter? (2)
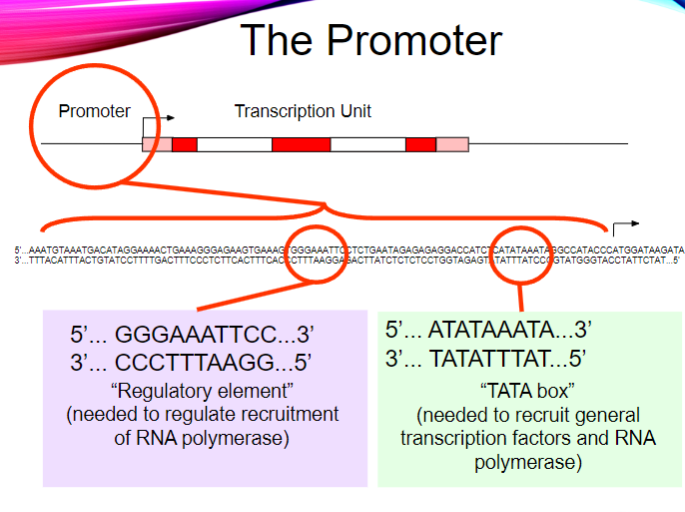
It regulates the recruitment of RNA polymerase.
It ensures proper transcription initiation.

What is the sequence of the TATA box? (1)
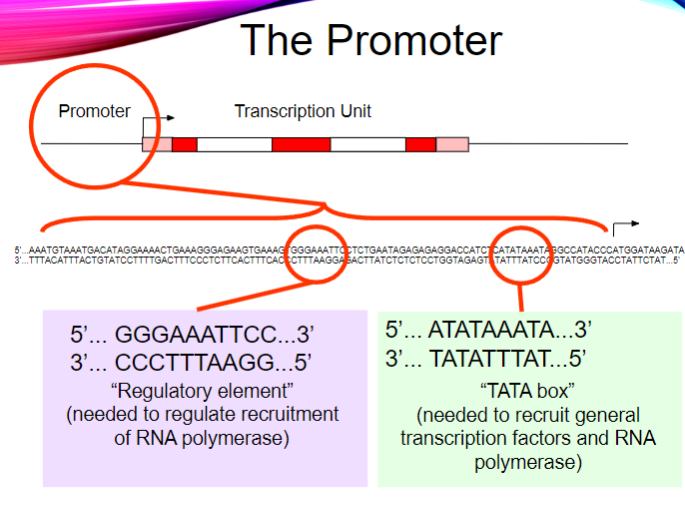
5' ... ATATAAATA ... 3'
3' ... TATATTTAT ... 5'
What is the function of the TATA box? (2)
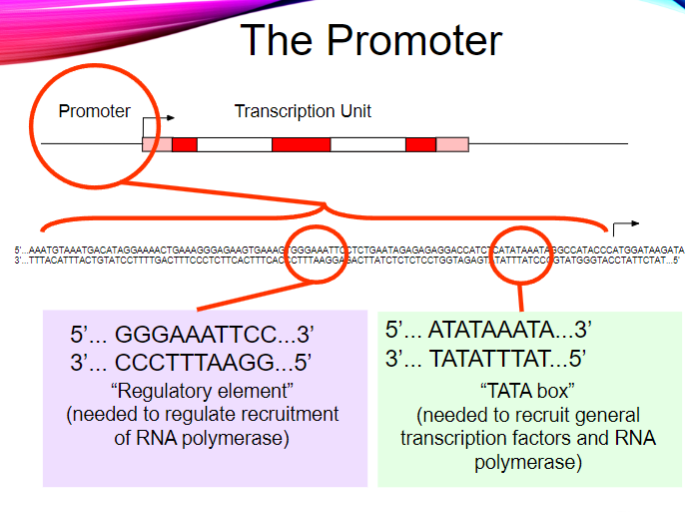
It recruits general transcription factors.
It helps recruit RNA polymerase for transcription initiation.
What is the main function of promoters? (1)
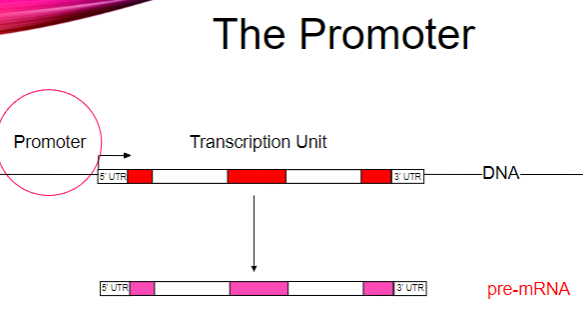
They recruit RNA polymerase to a DNA template.
How does RNA polymerase bind to the DNA template? (2)
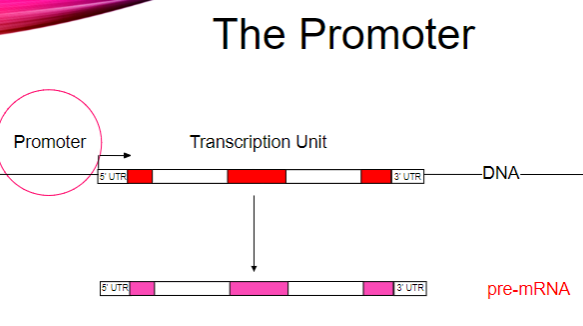
RNA polymerase binds asymmetrically.
It can only move in the 5’ to 3’ direction.
How is the binding of RNA polymerase regulated? (2)
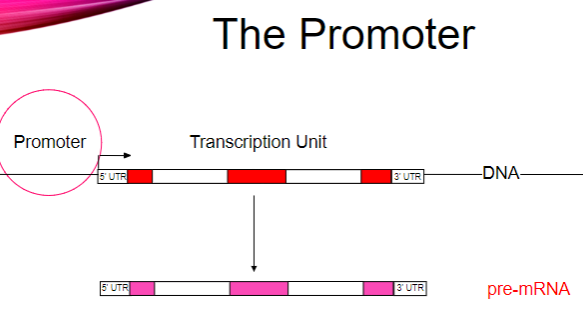
Binding regulation occurs via transcription factors.
Transcription factors assist in the recruitment of RNA polymerase to the promoter.
What is the function of enhancers in gene expression? (1)
Enhancers upregulate gene expression.
Where can enhancers be located? (2)
Enhancers can be found within the gene or many kilobases distant from it.
They are targets for transcription factors (activators).
What is the function of silencers in gene expression? (1)
Silencers downregulate gene expression.
How do silencers behave in relation to their location? (2)
Silencers are position-independent.
They also act as targets for transcription factors (repressors).
What is the role of insulators in gene regulation? (2)
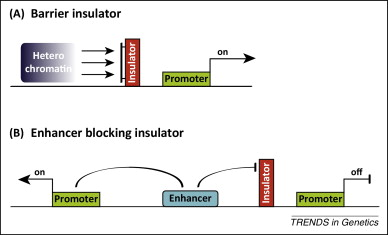
Insulators prevent enhancers and silencers from influencing other genes.
They act as short sequences that provide boundaries between regulatory elements.
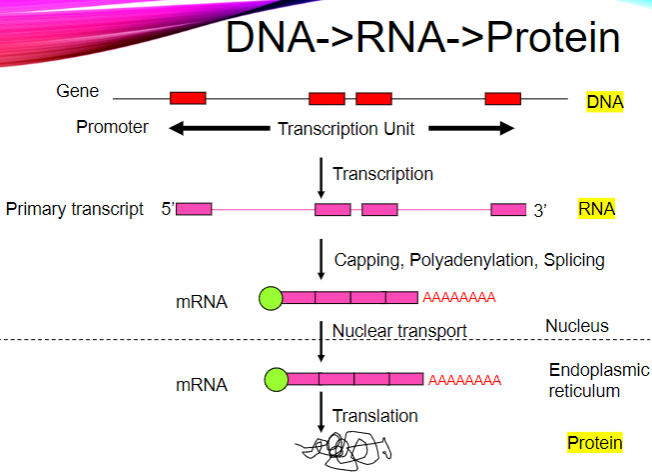
What catalyzes messenger RNA synthesis (transcription)? (1)
RNA Polymerase II.
In which direction does RNA Polymerase II transcribe? (1)
RNA Polymerase II transcribes in the 5’ to 3’ direction.
What does RNA Polymerase II transcribe after the transcription start site? (2)
RNA Polymerase II transcribes everything after the transcription start site, including exons and introns.
mRNA is post-transcriptionally modified.
How does RNA Polymerase II recognize promoters? (2)
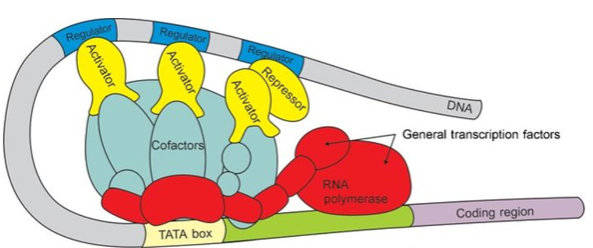
RNA Polymerase II recognizes promoters through the interaction with transcription factors such as TATA-binding protein (TBP), which binds to the TATA box in the promoter region.
General transcription factors (GTFs), including TFIID, TFIIB, TFIIE, and TFIIH, assemble at the promoter to facilitate the binding of RNA Polymerase II and the precise initiation of transcription.
Picture demonstrating mature mRNA synthesis
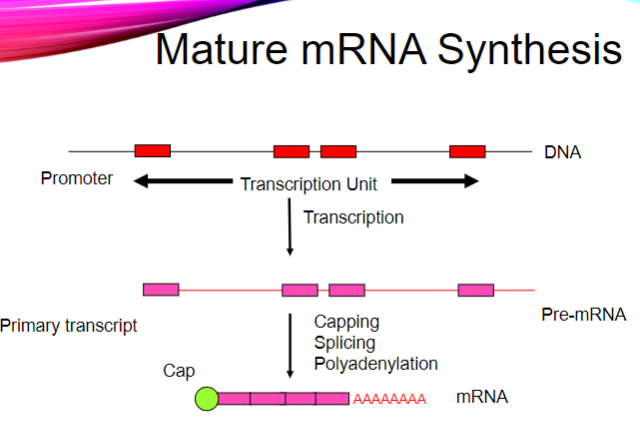
What are the three main post-transcriptional modifications of mRNA? (3)
Capped at the 5’ end.
Introns are spliced out.
Polyadenylated at the 3’ end.
What happens at the 5’ end of mRNA after 25-30 bases are added? (2)
A methylated cap is added to the 5’ end.
This cap is crucial for mRNA stability and recognition.
Which enzymes are involved in adding the 5’ cap to mRNA? (3)
RNA 5’-triphosphatase.
Guanylyltransferase.
N7G-methyltransferase.
What is the role of the bifunctional capping enzyme (CE) in capping? (1)
The bifunctional capping enzyme (CE) plays a key role in the capping of pre-mRNA by catalysing two essential steps:
-It adds a guanosine monophosphate (GMP) to the 5’ end of the mRNA, forming a 5’-5’ triphosphate bridge
-It methylates the guanosine cap at the N7 position, which is critical for mRNA stability and recognition by the ribosome during translation.
What other factor is required for the capping process? (1)
RNA Polymerase II is also required for the capping process.
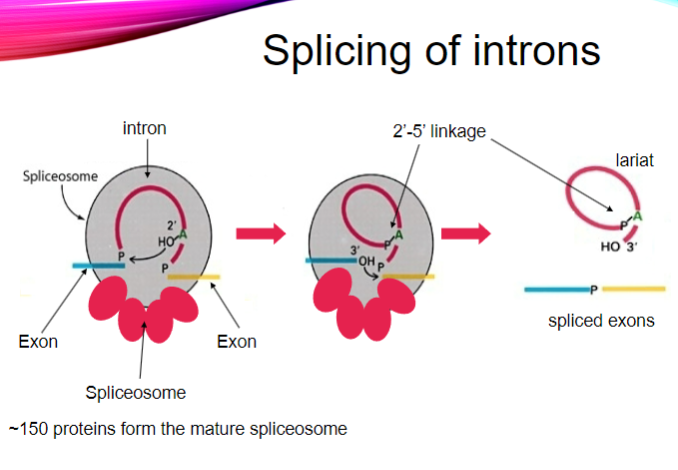
Picture demonstrating the splicing of introns
What is the role of CPSF (Cleavage and Polyadenylation Stimulating Factor) in mRNA processing? (2)
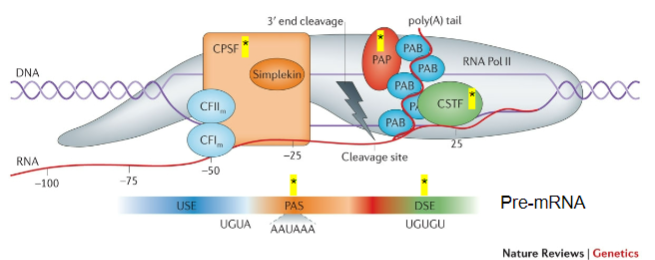
CPSF recognizes the Polyadenylation signal (PAS).
It acts on the cleavage site to facilitate polyadenylation.
What does CstF (Cleavage Stimulating Factor) recognize during the poly-A tail addition? (1)
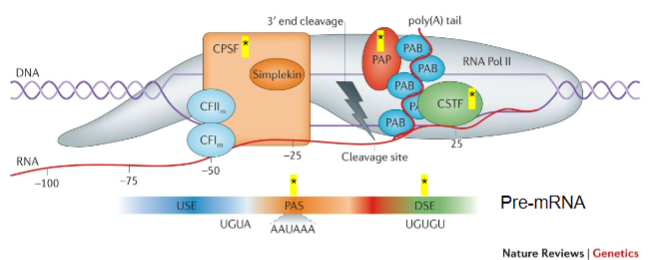
CstF recognizes GU-rich Downstream Elements (DSE).
What is the function of PAP (Poly-A polymerase) in the addition of the poly-A tail? (1)
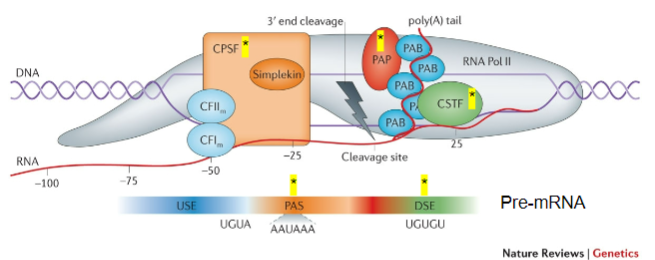
PAP is recruited to the cleavage site and adds multiple adenine (A) bases after cleavage.
What is the role of Poly-A Binding Protein (PAB) in the poly-A tail process? (1)
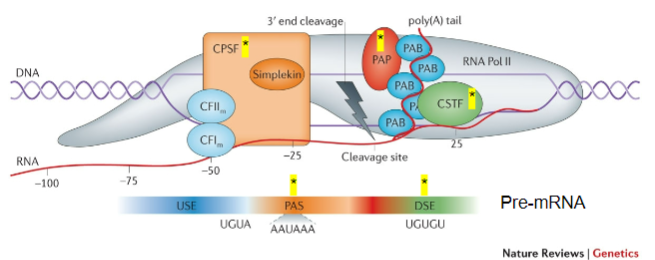
PAB binds to the poly-A tail and is involved in the stabilization and regulation of the mRNA.
Which other proteins are involved in the process of adding the poly-A tail? (3)
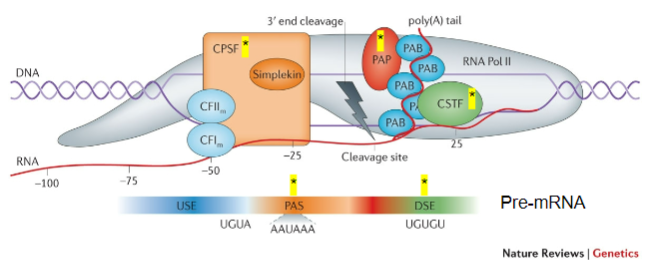
CFIm (Cleavage Factor Im).
CFIIm.
Simplekin.
Picture demonstrating Translation
What is the point of splicing in mRNA processing? (2)
To remove introns from the pre-mRNA.
To join exons together to form a continuous coding sequence.
What is alternative splicing? (2)
A process where exons can be skipped or included in the final mRNA.
This leads to the production of multiple protein variants (isoforms) from a single gene.
What are protein isoforms? (1)
Variants of a protein that arise from alternative splicing of a single gene.
How is DNA arranged in the cell generally? (2)
DNA is arranged non-randomly.
Most of the time, DNA is not organized into chromosomes.
How has the organization of DNA been determined? (2)
Using Hi-C, which detects genomic DNA sequences in close proximity.
Through high-throughput microscopy techniques.
What roles do the CTCF protein and Cohesin protein complex play in the genome? (2)
They help organize the genome into 3D structures.
They assist in bringing transcription machinery to specific regions of DNA.
What are the two compartments of the genome of a CELL? (2)
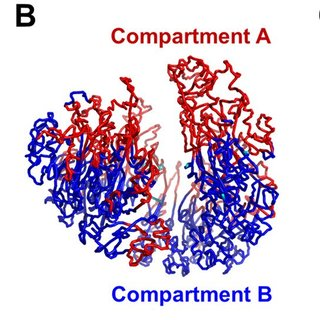
Compartment A: transcriptionally active with activating histone modifications.
Compartment B: transcriptionally repressed with repressive histone modifications.
How are the genome compartments distributed in the genome? (1)
The compartments are interspersed throughout the 2D sequence but brought close together in 3D space.
What are Topologically-Associated Domains (TADs)? (2)
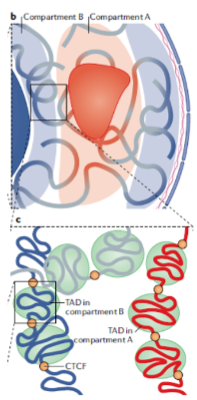
They are sub-compartments within individual genome compartments.
TADs are characterized by regions that interact more frequently with each other than with regions outside the domain.
What typically separates individual TADs? (2)
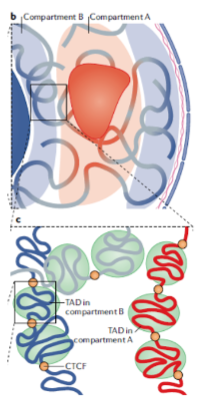
The Transcriptional Repressor CTCF protein.
CTCF acts as a boundary element, preventing interactions between TADs.
How do TADs contribute to genome organization? (1)
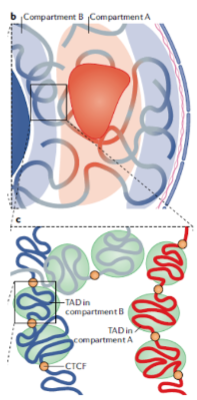
They help organize the genome into functional units, facilitating gene regulation and expression.
What is the role of the CTCF/Cohesin complex in transcription control? (2)
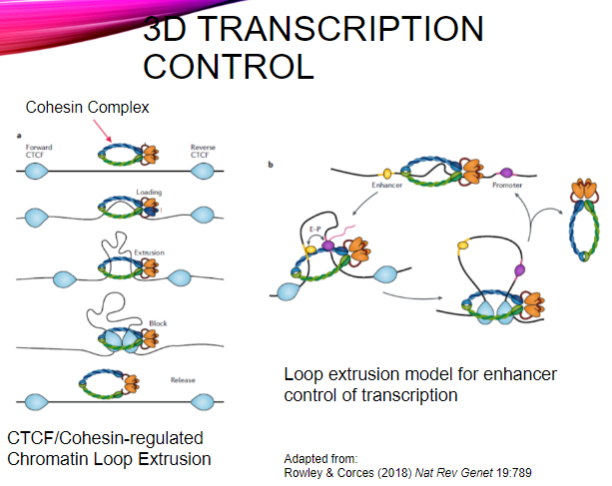
It regulates chromatin loop extrusion, which organizes DNA in three-dimensional space.
This organization facilitates the interaction between enhancers and their target promoters during transcription.
What is loop extrusion in the context of enhancer control? (2)
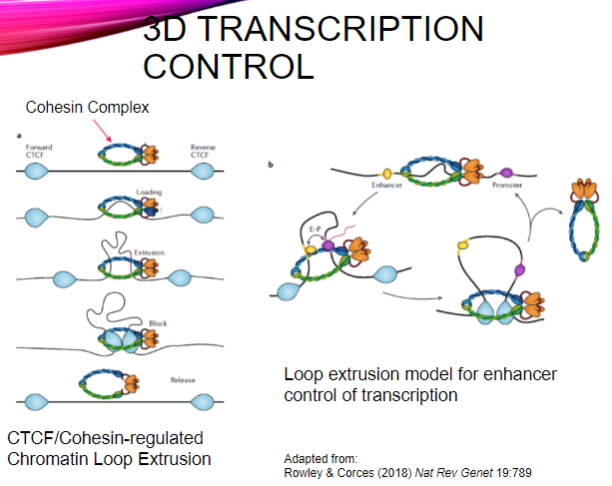
Loop extrusion is a mechanism by which the cohesin complex forms loops of DNA, bringing distant regulatory elements (enhancers) close to their target genes.
This process enhances the efficiency of transcription by promoting interaction between transcription factors and RNA polymerase at the gene's promoter.
How does the loop extrusion model contribute to transcription regulation? (1)
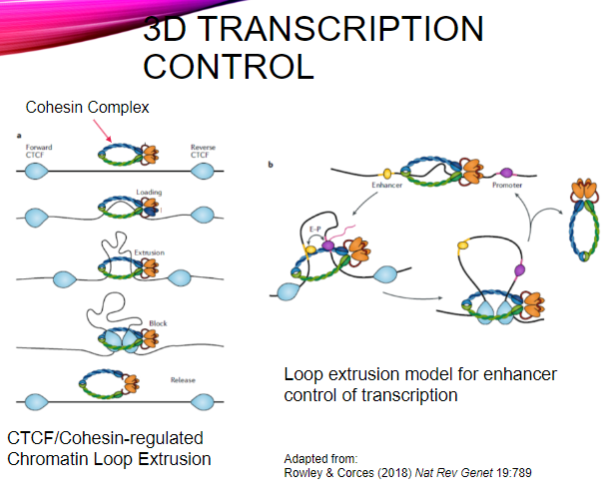
It allows for dynamic regulation of gene expression by enabling enhancers to interact with specific promoters, depending on the spatial arrangement of the chromatin.
What is the importance of CPSF, CstF, PAP, PAB, CFIm, CFIIm, and Simplekin (5)
CPSF: Key to recognizing the polyadenylation signal (PAS) and ensuring cleavage occurs at the correct site.
CstF: Recognizes GU-rich downstream elements and ensures the correct cleavage of the pre-mRNA, facilitating the recruitment of PAP.
PAP: Adds the poly-A tail, which is vital for mRNA stability, nuclear export, and translation efficiency.
PAB: Stabilizes the poly-A tail, preventing mRNA degradation and contributing to the regulation of mRNA lifespan.
Additional Factors (CFIm, CFIIm, Simplekin): They contribute to the overall regulation and coordination of the polyadenylation process, ensuring the efficiency and accuracy of mRNA maturation.