What is restriction mapping? (1)
Answer: Restriction mapping is a technique used to determine the positions of restriction enzyme cut sites within a DNA molecule. It helps in creating a map of the DNA structure based on fragment sizes produced by enzyme digestion.
What are restriction enzymes and their importance in restriction mapping? (2)
Answer: Restriction enzymes are proteins that cut DNA at specific recognition sequences. They are essential in restriction mapping as they generate DNA fragments that can be analyzed to create a map of the DNA sequence.
What are examples of recombinant proteins produced through genetic engineering? (3)
Answer: Examples of recombinant proteins produced through genetic engineering include:
-Insulin
-Interferon
-G-CSF.
What are transgenic organisms and their applications in genetic engineering? (2)
Transgenic organisms are those that have been genetically modified to carry genes from another species. They are used as disease models and for improving agricultural yields
What topics are covered in the DNA Technology Lectures? (4)
The DNA Technology Lectures cover the following topics:
DNA structure, denaturation & hybridisation
PCR amplification
Restriction, ligation & other enzymes
Cloning vectors
What enzymes are discussed in DNA Technology Lectures? (5)
The enzymes discussed include:
-Nucleases
-Ligase
-Polymerases
-Phosphatase
-Kinase
What do nucleases do? (3)
Nucleases degrade nucleic acids by hydrolysing phosphodiester bonds. They are classified into:
Ribonuclease (RNase): degrades RNA.
Deoxyribonuclease (DNase): degrades DNA.
They may also act as exonucleases (degrading from the end) or endonucleases (cleaving within the nucleotide chain).
What are restriction endonucleases (1), and what are their two functions? (2)
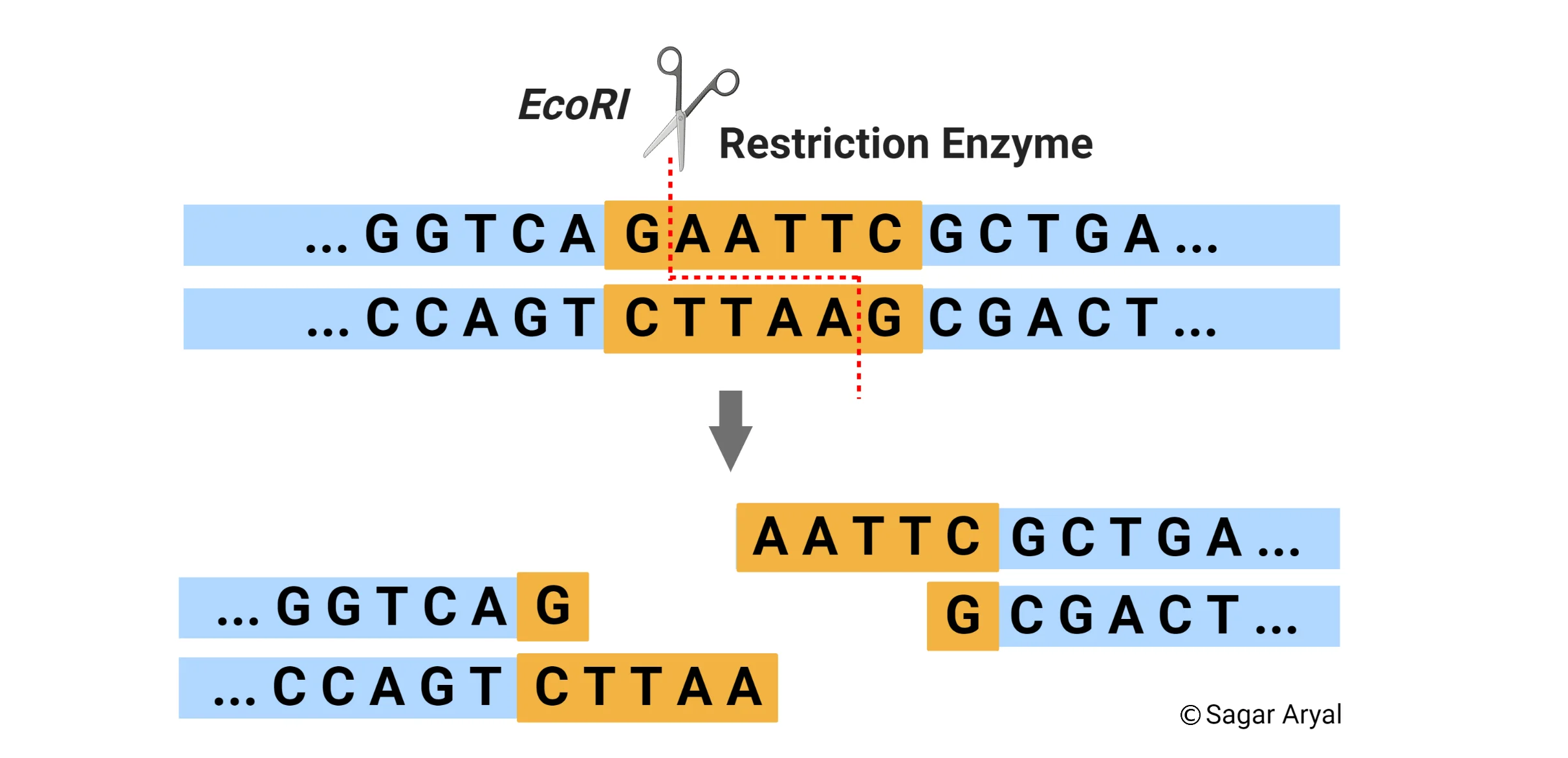
Restriction endonucleases are enzymes produced by bacteria to protect themselves from bacteriophage infections. These enzymes cut foreign DNA at specific recognition sequences, preventing the phage from replicating.
Their two functions are:
Recognizing specific DNA sequences: They identify unique short sequences of DNA.
Cutting the DNA at these sequences: They cleave the DNA at or near the recognition site.
What do different restriction enzymes recognize, and how long are the recognition sites? (2)
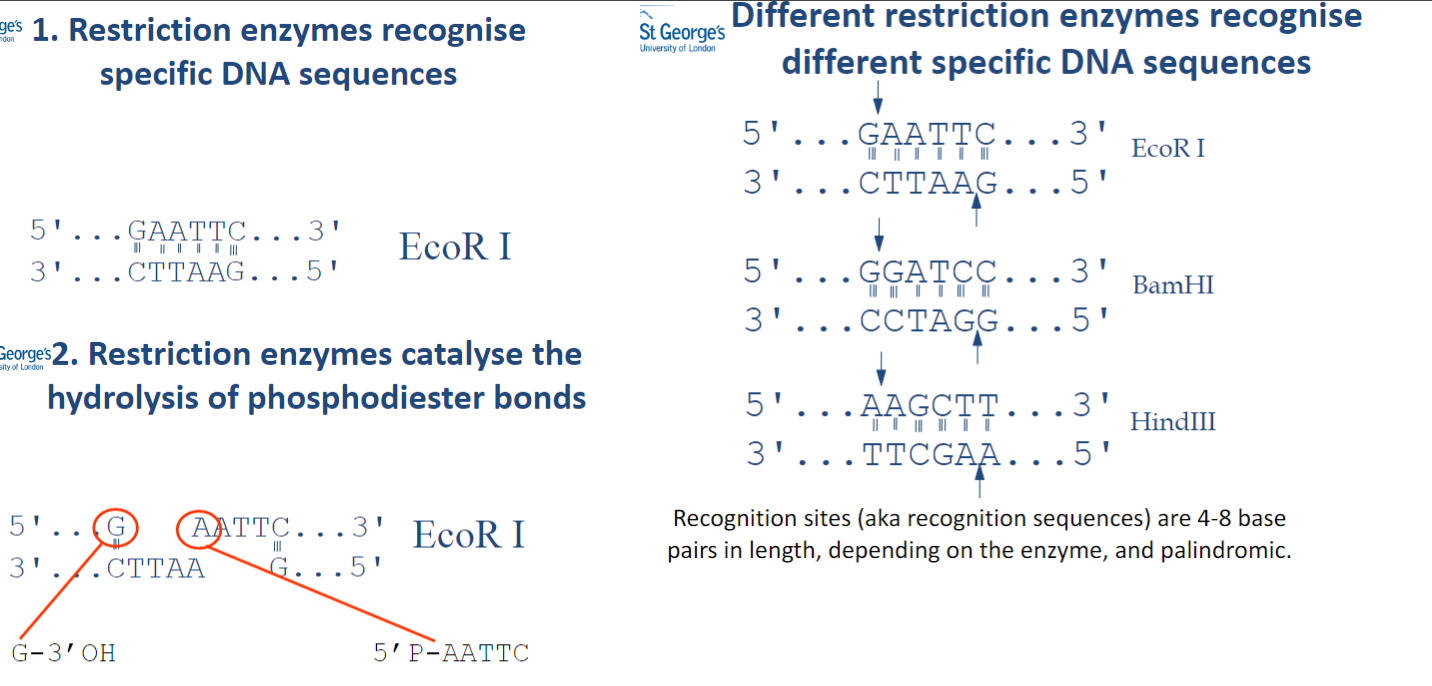
Different restriction enzymes recognize specific DNA sequences.
The recognition sites are 4-8 base pairs in length and are palindromic.
What do different restriction enzymes recognize, and how long are the recognition sites? (2)
Different restriction enzymes recognize specific DNA sequences.
The recognition sites are 4-8 base pairs in length and are palindromic.
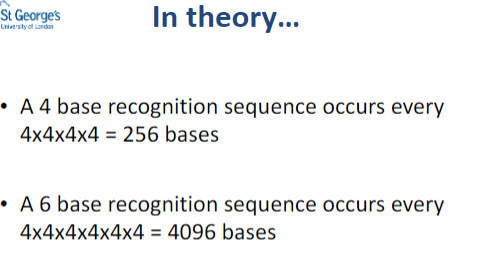
In theory.......
What are the two types of ends produced by nucleases? (2)
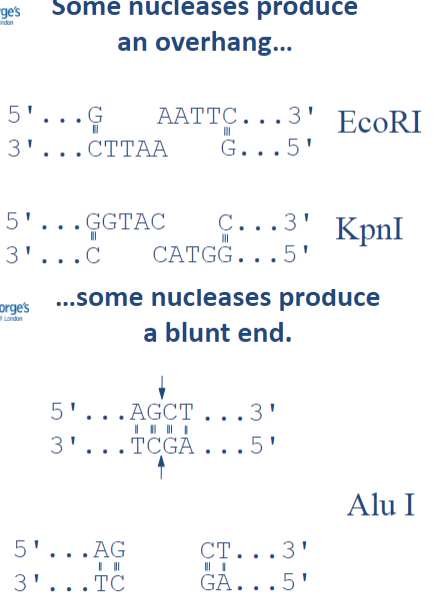
Some nucleases produce an overhang.
Some nucleases produce a blunt end.
What are restriction enzymes crucial for? (3)
Cloning: They cut DNA at specific sites, allowing insertion of foreign DNA into vectors (e.g., plasmids) for replication or expression.
Molecular Diagnostics: Used in techniques like RFLP analysis to detect mutations or variations in DNA sequences by cutting DNA at specific sites and observing fragment patterns.
Characterization of Plasmids: These enzymes cut plasmids at known sites to analyse their size and composition, helping researchers confirm the presence of desired DNA inserts.
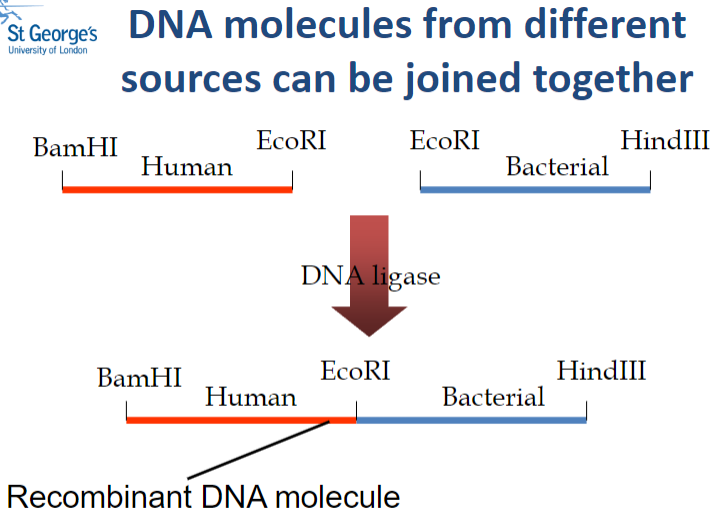
Picture demonstrating how DNA molecules from different sources can be joined together:
What role do restriction enzymes play in molecular diagnostics, specifically for sickle cell anaemia? (2)
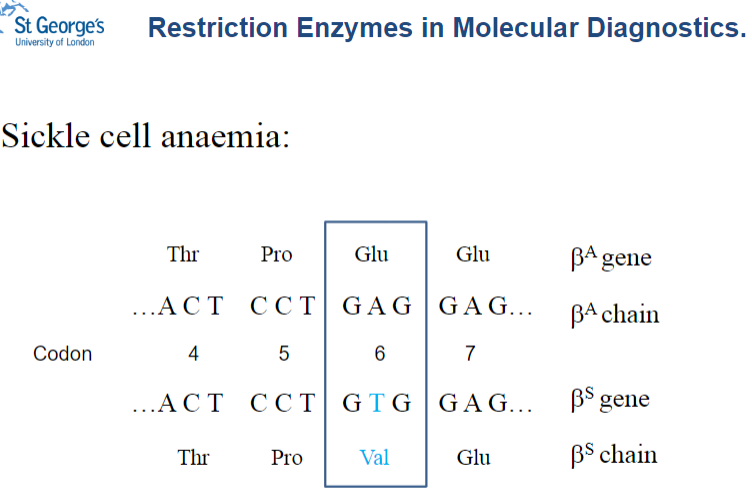
Restriction enzymes can be used to identify mutations responsible for sickle cell anaemia.
They recognize and cut at specific sites in the DNA, allowing detection of the sickle cell mutation through the pattern of DNA fragments.
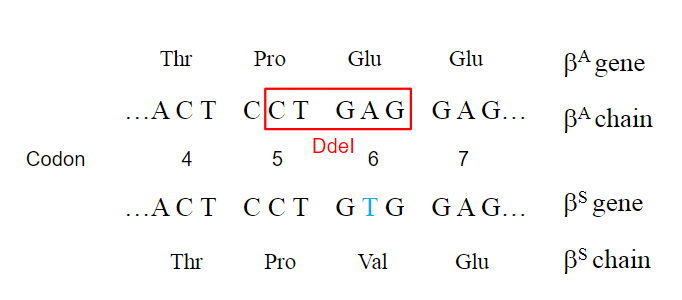
How does a single nucleotide change affect restriction enzyme sites in sickle cell anemia? (2)
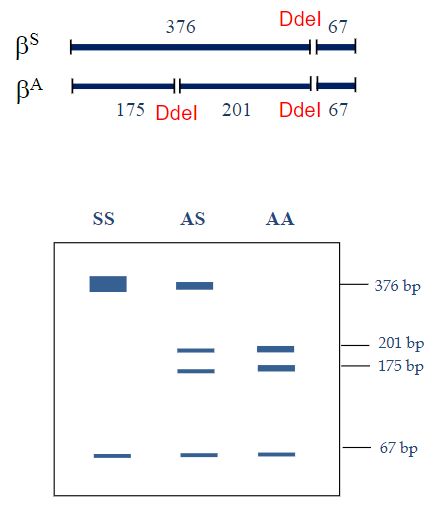
A single nucleotide change can create or destroy restriction enzyme recognition sites.
In sickle cell anaemia, the DdeI restriction enzyme site (5’CTNAG3’) is lost due to the mutation.
What are restriction maps, and why are they useful? (2)
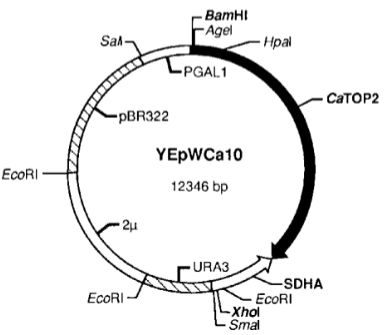
Restriction maps are diagrams that show the locations of restriction enzyme sites within a DNA molecule.
They provide a crude way of mapping an unknown molecule and are particularly useful for describing plasmids.
Picture demonstrating Restriction Mapping Example - Plasmid:
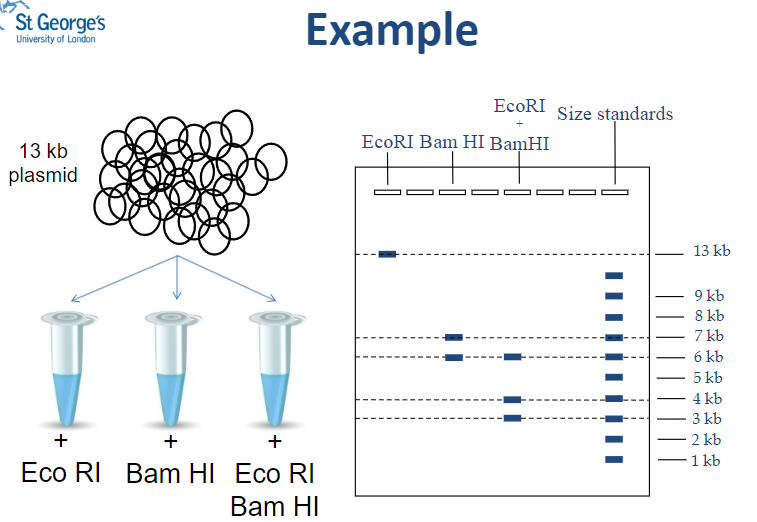
What is the total size of fragments produced when pDTL3 is digested with Bam HI? (1)
The fragments add up to 13 kb (7 kb + 6 kb).
How many times does Bam HI cut the plasmid pDTL3? (1)
Bam HI cuts the plasmid once, producing two fragments.
What is the length of the fragment produced when pDTL3 is digested with Eco RI? (1)
The length of the linear fragment is 13 kb.
How many times does Eco RI cut the plasmid pDTL3? (1)
Eco RI does not cut the plasmid, resulting in one linear fragment.
What is the total size of the fragments resulting from double digestion of pDTL3? (1)
The fragments add up to 13 kb (6 kb + 4 kb + 3 kb).
What is the biggest fragment resulting from double digestion of pDTL3? (1)
The biggest fragment is 6 kb.
Which enzyme gives a 6 kb fragment when pDTL3 is digested? (1)
The Bam HI enzyme gives a 6 kb fragment.
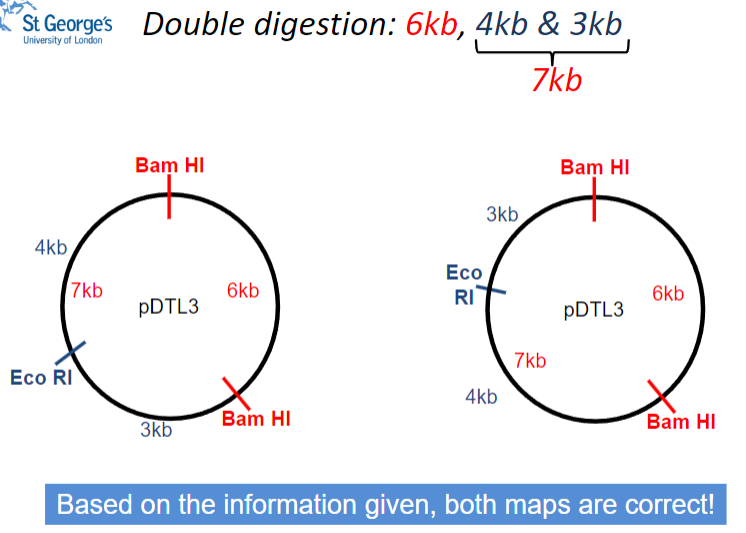
Picture demonstrating double digestion:
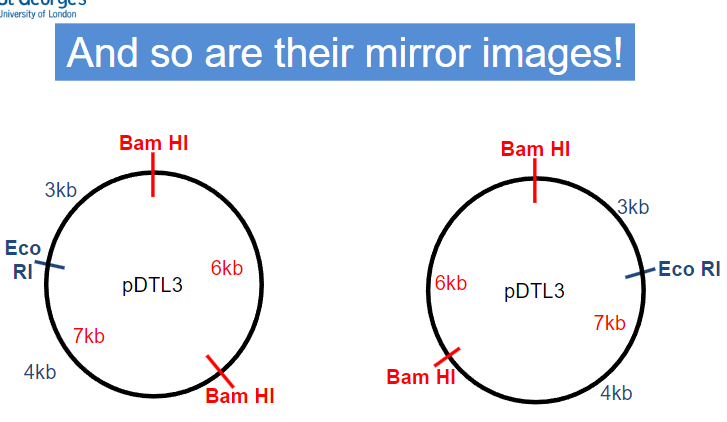
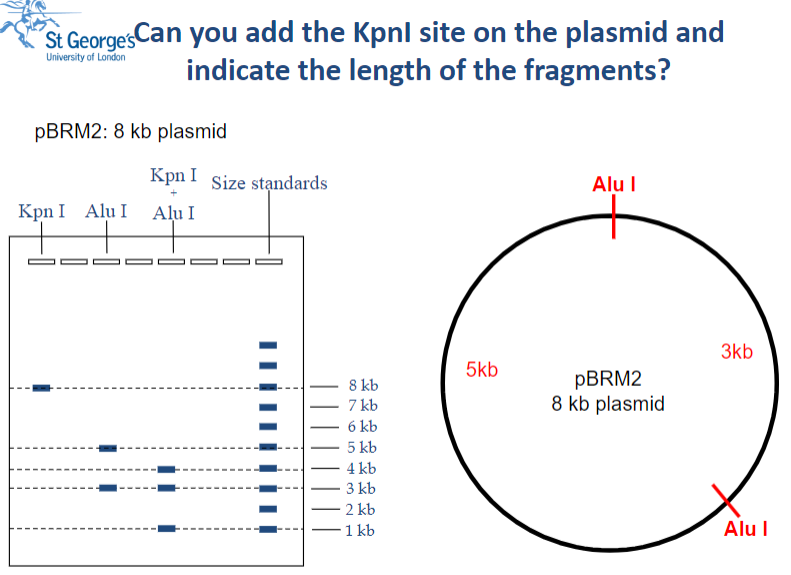
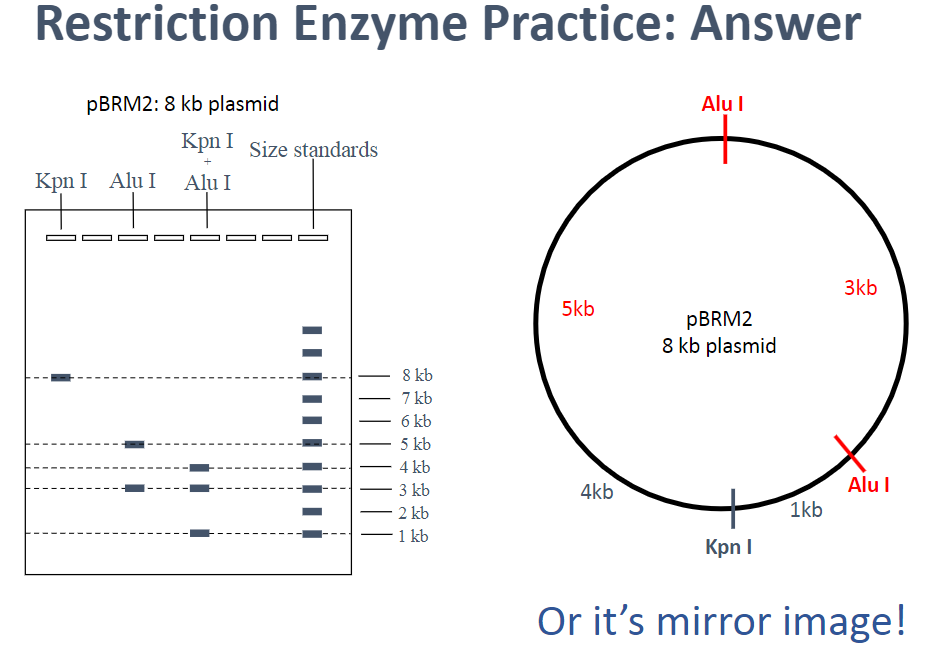
Restriction Mapping: Practice
What is DNA Ligase and what is its function? (2)
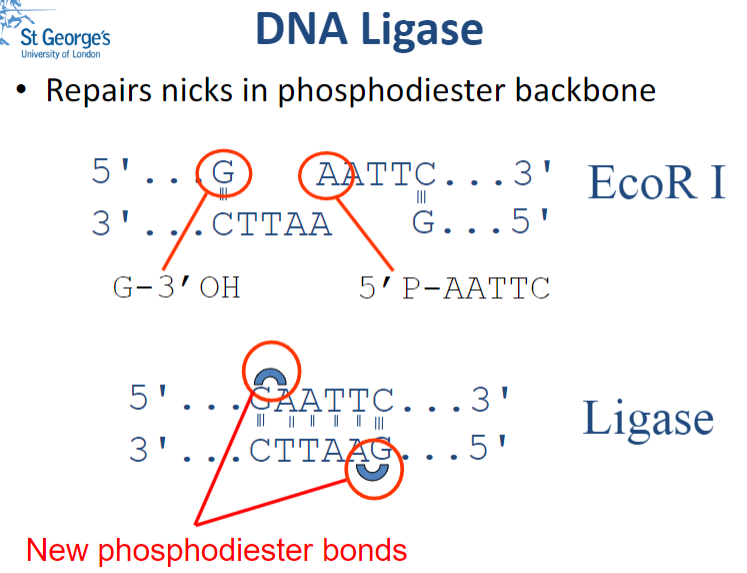
DNA Ligase is an enzyme that repairs nicks (small breaks) in the phosphodiester backbone of DNA by catalysing the formation of phosphodiester bonds.
Picture demonstrating the action of DNA Polymerase:
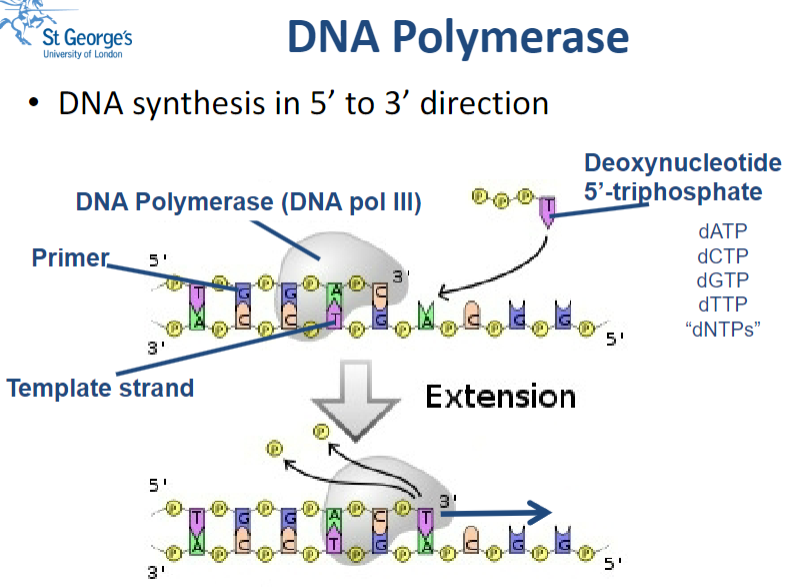
Why is DNA Polymerase used in molecular biology? (3)
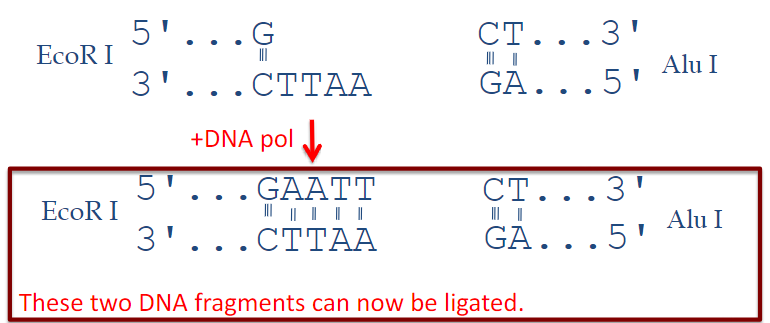
PCR amplification: DNA Polymerase is essential for amplifying specific segments of DNA through the polymerase chain reaction (PCR).
Generation of probes: It is used to synthesize DNA probes that can hybridize with target sequences.
Blunt-ending of DNA overhangs: DNA Polymerase can create blunt ends by filling in or chewing back overhanging nucleotides.
What is the function of Phosphatase in molecular biology? (2)
Hydrolyzes a phosphate group: Phosphatase removes phosphate groups from its substrates, which can be crucial for various biochemical processes.
Types of Phosphatase: Common examples include Calf Intestinal Alkaline Phosphatase and Shrimp Alkaline Phosphatase.
Why use a Phosphatase? (1)
To prevent cut plasmids from resealing.
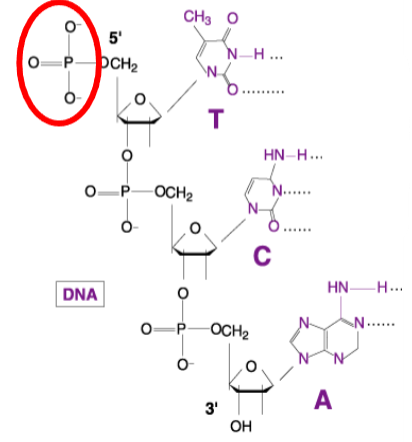
What is the function of Polynucleotide Kinase in molecular biology? (3)
Kinase Activity: Polynucleotide Kinase transfers a phosphate group from ATP to a substrate.
Phosphorylation: It specifically adds a phosphate to the 5' hydroxyl group of DNA or RNA.
Reaction Equation: The reaction can be summarized as:ATP+Substrate→(using kinase)→ADP+Phosphorylated-substrate
Why is Polynucleotide Kinase used in molecular biology? (3)
Phosphorylation: To phosphorylate chemically synthesized DNA, enabling it to be ligated to another fragment.
Sensitive Labeling: To label DNA sensitively for tracking purposes.
Labeling Methods: This can be achieved using:
Radioactively labeled ATP
Fluorescently labeled ATP
What are probes in molecular biology? (3)
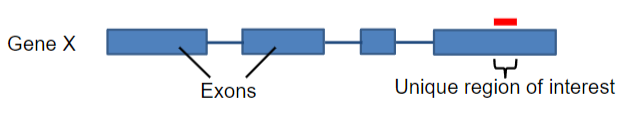
Definition: Probes are fragments of single-stranded DNA (ssDNA) or RNA.
Length: They typically range from 20 to 1000 bases in length.
Function: Probes are complementary to the gene of interest, allowing for specific detection and hybridization.
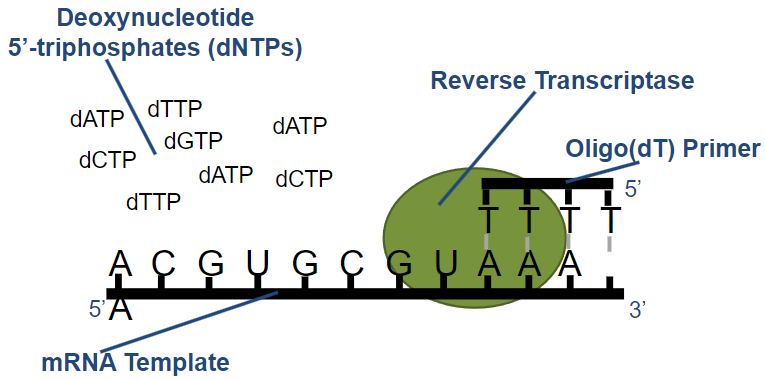
What is Reverse Transcriptase and its function? (3)
Definition: Reverse transcriptase is an RNA-dependent DNA polymerase.
Source: It is isolated from RNA-containing retroviruses.
Function: It synthesizes a DNA molecule complementary to an mRNA template using deoxynucleotide triphosphates (dNTPs).
What is Reverse Transcriptase? (1)
Definition: Reverse transcriptase is an RNA-dependent DNA polymerase.
What are the two types of primers used in reverse transcription (2), and what are their characteristics? (4)
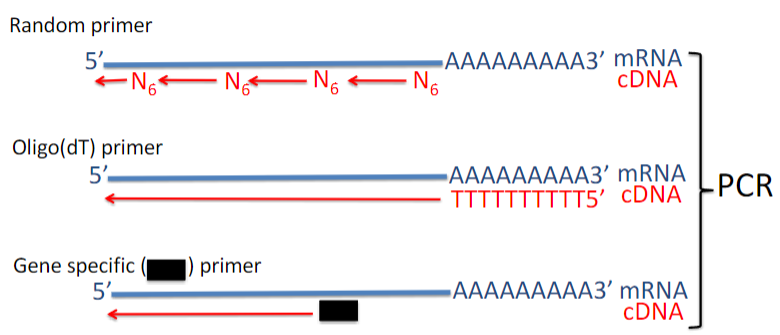
Random Primers:
Produce cDNAs up to 700 bp.
Cover the length of all RNA molecules.
Oligo(dT) Primers:
Useful for cloning cDNAs and creating cDNA libraries.
Some cDNAs generated may not be full-length.