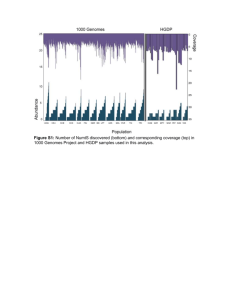
Ongoing DNA transfer from the mitochondrial to the
advertisement

Ongoing DNA transfer from the mitochondrial to the nuclear genome Bioinformatic and Comparative Genome Analysis Course HKU-Pasteur Research Center, Hong Kong, China August 17-29, 2009 Miria Ricchetti (mricch@ pasteur.fr) QuickTime™ and a decompressor are needed to see this picture. 1. The genomes of eukaryote cells The genomes of eukaryote cells mitochondria chloroplasts (in plants) nucleus Origin of mitochondria and chloroplasts Endosymbiont theory Mitochondria and chloroplasts are relics of free-living bacteria that formed a symbiotic association with the precursor of the eukaryotic cell Origin of mitochondria and chloroplasts Animals Fungi Plants Slime moulds Protozoa Algae Chloroplasts Sulfolobus EUCARYOTES Thermoplasma Extreme halophiles Cyanobacteria Mitochondria Methanogens Purple photosynthetic bacteria Thermoacidophiles Gram-positive bacteria Spirochetes Green photosynthetic bacteria EUBACTERIA ARCHAEBACTERIA UNIVERSAL ANCESTOR Mitochondria Mitochondria generate most of the cell's supply of adenosine triphosphate (ATP), used as a source of chemical energy. Mitochondria are also involved in other processes, such as signaling, cellular differentiation, cell death, as well as the control of the cell cycle and cell growth. Mitochondria have been implicated in several human diseases, including mitochondrial disorders and cardiac dysfunction, and may play a role in the aging process. View of mitochondria within a cell McBride HM et al. 2006. Curr. Biol. 16: R551-600;Bossy-Wetzel E, et al 2003. Curr Opin Cell Biol 15, 706-16.. Chloroplasts Chloroplasts, also called plastids, are organelles found in plant cells and other eukaryotic organisms that conduct photosynthesis. During photosynthesis, chloroplasts capture light energy to conserve free energy in the form of ATP and reduce NADP to NADPH. Eberhard S. et al. 2008. Ann. Rev. Geneti. 42 463-515 Puthiyaveetil S, Allen JF. 2009, Proc Biol Sci. 276:2133-45 Genomes size Saccharomyces cerevisiae Homo sapiens Nuclear genome Mitochondrial genome 1.2 x 107 bp 85,779 bp 2.9 x 109 bp 16,554 bp Mt genome size size mt genome (kbp) Reclinomonas americana (protist) Arabidopsis thaliana Saccharomyces cerevisiae 69 67 367 @ 45 85.8 Plasmodium falciparum (parasitic protist) 17 12-24 average Homo sapiens and most vertebrates protein-coding genes 16.5 13 6 3 Smallest free-living a-proteobacterial genome Bartonella henselae < 2000 @1600 Mitochondrial Genome Products The mitochondrial genome: • encodes some proteins but most proteins for organelle function are encoded by chromosomal genes and imported from cytoplasm • encodes most of RNA components of protein synthesis (mRNA, rRNA, tRNA) Genes coding for mitochondrial function and propagation in Saccharomyces cerevisiae mt proteins are coded: about 400 in the nucleus (200 are of bacterial origin) 17 in the mt Mitochondrial genome products (in humans) Protein Complex Encoded by Encoded by Mitochondrial Nuclear Genome Genome __________________________________________________________________ Oxidative phosphorylation NADH dehydrogenase Succinate CoQ reductase Cytochrome b-c1 complex Cytochrome c oxidase complex ATP synthase complex 7 subunits 0 subunits 1 subunit 3 subunits 2 subunits >41 subunits 4 subunits 10 subunits 10 subunits 14 subunits none none Ribosomal proteins 2 rRNAs 22 tRNAs (13 mRNAs) none Other mitochondrial proteins none All, e.g., mitochondrial DNA pol, RNA pol, other enzymes, structural proteins Protein synthesis apparatus (~80 in total) Mitochondrial Genome • Circular • Much smaller than nuclear genome with a compact genetic organization (human 16.5 kb, yeast 85 kb, melon 2500 kb) • in humans: • ~ 10 identical molecules per mitochondrion (100s-10,000s mitochondria per cell) • 37 genes ; no introns (but present in S. cerevisiae) • 13 are protein-coding genes (respiratory complex) • 24 are non-coding RNA genes 2 ribosomal RNAs, 22 transfer RNAs Genetic Organization of Human Mitochondrial DNA H strand synthesis D-loop H strand transcription 16S rRNA 23S rRNA CYB L strand transcription H STRAND ND1 ND6 16.5 kb ND5 ND2 L strand synthesis L ND4 STRAND CO1 ND4L ND3 CO2 CO3 Genes encoding proteins rRNA genes tRNA genes Genetic organization of S. cerevisiae mitochondrial DNA Exons 21S rRNA CO2 var Noncoding ATPase 9 Cytochrome b Introns CO3 85.7 kb par ATPase 6 ATPase 8 15S rRNA CO1 Mitochondria and human diseases Mitochondrial dysfunctions are responsible for several human diseases, in particular neurodegenerative disorders. This is the result of of the mitochondria's central role in energy production, reactive oxygen species (ROS) biology, and apoptosis. Mitochondrial DNA, whose integrity is strictly related to that of the mitochondrion itself, appears to be involved in pathologies and in the process of ageing. mtDNA mutations are sufficient by themselves to generate major clinical phenotypes Bossy-Wetzel E et al. 2003. Curr Opin Cell Biol 15, 706-16. Scott SV, et al. 2003. Curr Opin Cell Biol 15, 482-8. Wallace DC & Fan W 2009 Genes Dev. 23:1714-36 Mitochondria and human diseases Also defects in mtDNA maintenance are associated with an increasing number of human diseases (i.e. optic atrophy) Spelbrink JN et al. 2001 Nat Genet, 28:223-31 Trifunovic A et al. 2004 Nature, 429:417-23 Alexander C et al. 2000 Nat Genet, 26:211-5. Delettre C et al. 2000 Nat Genet, 26:207-10 Mitochondria and human diseases Some diseases are associated with the mutagenic insertion of mt DNA in the nuclear genome. (it will be discussed later) Turner C et al. 2003. Hum Genet 112, 303-9. Borensztajn K et al. 2002. Br J Haematol 117, 168-71 Goldin E et al. 2004. Hum Mutat. 24, 460-5. Willett-Brozick JE et al. 2001. Hum Genet 109, 216-23. Chloroplast genome size protein-coding genes Porphyra purpurea (red alga) 253 average 130 group of Apicomplexans (parasitic protists) ancestral cyanobacterial genome 30 @ 3000 Genome size reduction in mitochondria • transfer to the nucleus • function substituted by unrelated nuclear coded proteins (multi subunit RNA pol single subunit RNA pol) • loss of function: ex. complex I (nad) in S. cerevisiae Successful gene transfer requires: • movement of nucleic acid from the mt to the nucleus • integration of the DNA in the nuclear genome • expression of the transfered gene (different codon usage) • protein must acquire a transit peptide to allow access to the organelle • coordination of the expression to different energy needs Phylogenetic trees constructed by nuclear and mt DNA, suggest that mt and nuclear genomes have evolved in concert throughout much, if not most, of the evolutionary history of the domain Eukarya. from: Gray et al, Genome Biology (2001) Why do organellar genes and up in the nucleus? Characteristics of the mt genome • haploid genome • relatively small size • maternal inheritance • normally does not undergo recombination • relatively rapid sequence evolution higher risk of genetic drift Some references and reviews on the endosymbiotic theory and on the origin of organelles Margulis, Lynn, 1970, Origin of Eukaryotic Cells, Yale University Press. Raven JA, Allen JF. 2003. Genomics and chloroplast evolution: What did cyanobacteria do for plants? Genome Biol. 4:209 Andersson SG, Karlberg O, Canback B, Kurland CG. 2003. On the origin of mitochondria: a genomics perspective. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 358:165–7–9 Keeling PJ, Palmer JD. 2008. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9:605–18 2. Intercompartemental DNA transfer Theoretical DNA transfer in eukaryote cells mitochondria chloroplasts (in plants) nucleus Chloroplast DNA transfer in the nucleus Chloroplast DNA transfer in the nucleus • entire genes (during evolution) • infA in angiosperms • 33 kb of cpDNA on chromosome 10L of rice • transfer of chloroplast DNA into the nuclear genome of Nicotiana tabacum (preintegrated neomycine phosphotransferase gene) frequency: 1:16,000 • chloroplast DNA fragments (NUPT), ongoing Millen et al, 2001 Plant Cell 13: 645; Yuan et al, 2002 Mol. Gen. Genet. 267: 713 Huang et al, 2003 Nature. 422: 472 Nuclear DNA transfer in mitochondria Nuclear DNA transfer in mitochondria • 1 possible case (intra or interspecies ?): gene homologue of bacterial MutS in the mt of the coral Sarcophyton glaucum • interspecies (horizontal transfer): ribosomal (rps2 and rps11) and respiratory (atp1) proteins between distantly related flowering plants Pont-Kingdon et al, 1998 J.Mol.Evol. 46: 419; Bergthorsson et al, 2003 Nature 424: 197 Chloroplast DNA transfer in mitochondria Chloroplast DNA transfer in mitochondria • tRNA set in higher plants • 12 Kbp of DNA (originated from the inverted repeated region of the chloroplast genome) in maize • 5 independent transfers of the chloroplast rbcL to the mt genome in angiosperms (rbcL :ribulose-1,5-biposphate carboxylase/oxygenase) • 17 stretches of plastid-like sequences found in the mt DNA of Oryza sativa L. (rice) 17-6653 bp; 6.3% of mt genome; 61-100% identity Ellis J. 1982. Nature 299:678–79 ; Cummings et al, 2003 Curr. Genet 43: 131; Stern & Lonsdale, 1982 Nature 299: 698; Notsu et al, 2002 Mol. Genet. Genomics 268: 434) Chloroplast DNA transfer in mitochondria and then in the nucleus ? In plants, a few (nuclear) genes that code for mt proteins are derived by duplication from nuclear genes of ancestrally chloroplast origin (rps13 in some angiosperms, ribosomal protein S13 in Arabidopsis thaliana) Adams et al, 2002 Plant Cell 14: 931; Mollier et al, 2002 Curr. Genet. 40: 405 Nuclear DNA transfer in chloroplasts Nuclear DNA transfer in chloroplasts • two open reading frames, int and dpoB, in the large inverted repeat of the chloroplast genome of the green alga Oedogonium cardiacum no sequence similarity with genes in chloroplast genomes Mt donor unkonwn Brouard JS et al. 2008. BMC Genomics 9:290 Mitochondrial DNA transfer in chloroplasts not found… until now Mitochondrial DNA transfer in the nucleus Mitochondrial DNA transfer in nucleus • entire genes (during evolution) • mt DNA frangments (NUMT), ongoing Mt DNA insertions in the nuclear genome • genes (ceased in animals, detected in plants) Ongoing gene transfer from mt to the nucleus (in plants) ex. rsp10 gene (protein of the mt ribosome) in angiosperms from: Knoop et al. 1995, Curr. Genet; Adams et al. 2000, Nature Ongoing gene transfer from mt to the nucleus (in plants) Nucleus Mitochondria SDH2 RPS14 RPL5 rice RPS14 RPL5 wheat RPS14 RPL5 maize RPL5 yRPS14 rice RPL5 yRPS14 wheat RPL5 yRPS14 maize see: Sandoval et al. 2004 Gene 324:139 Dual expression (mt and nucleus) of atp9 in Neurospora The nuclear and the mt copy of atp9 are expressed at different stages of the life cycle (germinating spores versus vegetative cells) Bittner-Eddy et al. 1994 J. Mol. Biol. 235: 881 Dual expression (mt and nucleus) of cox2 in legumes Adams et al. 1999 PNAS 24:13863 Mt insertions in the nuclear genome • genes (ceased in animals, detected in plants) • 620 kbp insertion in Arabidopsis thaliana Large mt DNA insertion in the centromeric region of chromosome 2 of Arabidopsis thaliana A D C B duplications D telomere A D A D A C B Mt DNA insertion (@620 kbp) from Stupar et al. 2001, PNAS 98, 5099. See also Lin et al. 1999, Nature 402, 761. centro mere Mt insertions in the nuclear genome • genes (ceased in animals, detected in plants) • 620 kbp insertion in Arabidopsis thaliana • 7.9 kbp in felines A 7.9 Kbp insertion of mt origin in the nuclear genome of domestic cat 7.9 kb of a typically 17.0-kb mitochondrial genome inserted to a specific nuclear chromosomal position in the domestic cat. the intergrated segment has subsequently become amplified 38-76 times and now occurs as a tandem repeat macrosatellite Lopez et al. 1994, J. Mol. Evol. 39, 554 Mt insertions in the nuclear genome • genes (ceased in animals, detected in plants) • 620 kbp insertion in Arabidopsis thaliana • 7.9 kbp in felines • DNA fragments of about 300 bp (NUMTs) till > 14 kbp (in most studied eukaryotes) Mitochondrial, nuclear and NUMT sizes in some eukaryotic genomes size mt DNA (bp) Homo sapiens 16.571 Mus musc ulus 16.299 Rattus norvegic us 16.3 Fugu rubripes 16.447 Caernorhabditis elegans 13.794 Drosophila melanogaster 19.517 Anopheles gambiae 15.363 Ciona intestinalis 14.788 Plasmodium falc iparum 5.697 Sac c haromyc es c erevisiae 85.779 sc hisac c haromyc es pombe 19.431 Arabidopsis thaliana 366.924 Oryza sativa 490.52 -4 nuc lear (Mbps) NUMTs (BLASTN threshold 10 ) 2910 279.17 2500 53.453 2800 6.34 320 5.624 97 126 122.7 534 278.2 0 116.7 11.51 22.9 152 12.5 1.241 12.5 1.614 115.4 198.105 420 409.104 from Richly & Leister 2004 Mol Biol Evol 21, 1081 % 10- 3 9,.6 2.1 0.2 1.8 0.1 0.4 0.0 9.9 0.7 9.9 12.9 171.7 97.4 NUMTs in six hemiascomycetous yeast species Yeast species S. cerevisiae C. glabrata K. thermotolerans K. lactis D. hansenii Y. lipolytica Nuclear genome size (Mb) 12.1 12.3 10.4 10.6 12.2 20.5 Mitochondrial genome size (kb) 85.7 20 23.5 40.2 29.4 47.9 NUMT number 32 14 1 8 145 47 Total transferred mitochondrial DNA (bp) 2356 1423 25 403 9377 2005 Transferred mitochondrial DNA (%) 2.7 7.1 0.1 1 31.8 4.2 from Sacerdot et al. 2008 FEMS Yeast 8, 846-850 Why variable abundance of NUMTs in different species ? • frequency of DNA transfer ? • vulnerability of mt to stress & other factors? • n° of mt/cell ? (i.e. Plasmodium) • n° of somatic cell divisions from zygotes to meiosis • efficiency of nuclear import of mt DNA and/or its integrattion into the nuclear genome • rate of loss of NUMTs ? • rates of DNA loss varies from fragments and among species • however, no NUMT loss has been show till now from Richly & Leister 2004 Mol Biol Evol 21, 1081 Types of NUMTs and NUPTs from Leister. 2006 Trends in Genetics 21, 655-663 3. How do NUMTs integrate in the nuclear genome? Mitochondrial DNA integrate in the nuclear genome during double-strand break (DSB) repair in yeast Ricchetti et al.1999 Nature, 402:96-100 Yu & Gabriel 1999 Mol. Cell 4: 873-881 Mt DNA insertion in the nuclear genome via Non-Homologous End-Joining (NHEJ) intact chromosome DSB + Ricchetti et al.1999, cited mt DNA Analysis of mt insertions during the repair of DSBs in yeast DSB NHEJ mt DNA insertion Mt insertions found at DSB sites in yeast Digested I-SceI sites: ATTACCCTGTTAT3' TAATGGGAC + CAGGGTAAT 3'TATTGTCCCATTA 34pAT9 ATTACCCTGTTATattattattttttattattaataataataatttatagggtttattctgttttatcataa atacgtaaatatctaacttagctctcaaattatattacTAACAGGGTAAT (A) 34pAS16 ATTACCCTGTTATttagaatatttttaattaaataatataattaaatgaataccaaacttatattatattta tatttatatttatatttcTAACAGGGTAAT (A) 34pAS15 ATTACCCTGTTATctttattatatttaagaatattattataattattattattattattatttttaataatt aaaaatattaataataagtaaatattaattattgttcatttaatcattccaaaaatttaggtaatgatactg cttcgatcttaattggcatatttgcatgacctgtcccacacaactcagaacatgctccggccacgggagccg gaaccccgaaaggaggaataagataaatatatagCAGGGTAAT (ATAA) (T) 622pBS8 ATTACCCTGTTAagtttccatagaagtaataataataataaatatattaaatattaatataattattaatta aaataactaatttagatcaatctaaaaaatctaagtgtttagatgataataaagaatatttattaaagtatt ctattactttaatattttTAACAGGGTAAT (A) Ricchetti et al.1999, cited NUMTs in the yeast nuclear genome Saccharomyces cerevisiae Nuclear genome Mitochondrial genome 1.2 x 107 bp 85,779 bp by DSB repair: 6 NUMTs size: single 47-97 bp multiple 166-382 bp homology :100% size: 22-230 bp homology :86-100% by BLAST search: 30 NUMTs Ricchetti et al.1999, cited Yeast mitochondrial genome and origin of NUMTs NUMTS at DSBs in the yeast genome found > once 4. Impact of NUMTs insertion on the eukaryotic genomes 211 NUMTs detected in humans • Sequence analysis of the genome of Homo sapiens • PCR sampling of humans from different ethnic backgrounds Ricchetti et al.2004 PLoS Biology, 2:96-1313 PCR amplification of NUMTs in the human genome NUMT amplified DNA fragment NUMT NUMT present NUMT absent Insertion polymorphism of three NUMTs Insertion polymorphism of NUMTs in humans 1 individual 2 sex 3 NUMT 1-74 4 NUMT 2-132 5 NUMT 2-53 6 NUMT 12-89 7 NUMT 13-75 8 NUMT 18-192 9 NUMT 11-541 Caucasian-1 Caucasian-2 Caucasian-3 Caucasian-4 Caucasian-5 Caucasian-6 Caucasian-7 Caucasian-8 Caucasian-9 Caucasian-10 African-1 African-2 African-3 Pygmy-1 (Biaka) Pygmy-2 (Mbuti) Pygmy-3 (Mbuti) Pygmy-4 (Biaka) Chinese-1 Chinese-2 Japanese-1 Japanese-2 Female Male Male Male Female Male Female Male Male Female Male Female Male Male Male Female Female Male Male Male Female -/-/-/+ -/-/+ -/-/-/-/-/+ +/+ -/+ -/+/+ -/-/-/-/+ -/-/-/- +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ -/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ -/+/+ -/+ -/+ -/+ -/+ -/+/+ -/+ -/+ -/+ -/+/+ -/+ +/+ -/-/+ -/-/+ -/+ -/+ -/+ -/-/-/-/-/-/-/+ -/+ +/+ -/-/-/+ -/-/+ -/+ -/-/-/+ -/-/- +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ -/+ -/+ +/+ -/-/-/+/+ -/-/-/-/+/+ -/+ +/+ -/+ -/+ -/-/-/+ -/-/+ -/-/+ -/- +/+ -/+/+ +/+ -/+ -/+ -/+ -/+/+ -/+ -/-/+ -/+ -/-/-/-/-/-/+ -/+ -/+ * */* * * Ricchetti et al.2004, cited * * * * * Frequency of alleles carrying the NUMT NUMT % 2-132 13-75 2-53 18-192 12-89 1-74 98 95 48 29 21 21 Insertion polymorphism of NUMTs in humans 1 individual 2 sex 3 NUMT 1-74 4 NUMT 2-132 5 NUMT 2-53 6 NUMT 12-89 7 NUMT 13-75 8 NUMT 18-192 9 NUMT 11-541 Caucasian-1 Caucasian-2 Caucasian-3 Caucasian-4 Caucasian-5 Caucasian-6 Caucasian-7 Caucasian-8 Caucasian-9 Caucasian-10 African-1 African-2 African-3 Pygmy-1 (Biaka) Pygmy-2 (Mbuti) Pygmy-3 (Mbuti) Pygmy-4 (Biaka) Chinese-1 Chinese-2 Japanese-1 Japanese-2 Female Male Male Male Female Male Female Male Male Female Male Female Male Male Male Female Female Male Male Male Female -/-/-/+ -/-/+ -/-/-/-/-/+ +/+ -/+ -/+/+ -/-/-/-/+ -/-/-/- +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ -/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ -/+/+ -/+ -/+ -/+ -/+ -/+/+ -/+ -/+ -/+ -/+/+ -/+ +/+ -/-/+ -/-/+ -/+ -/+ -/+ -/-/-/-/-/-/-/+ -/+ +/+ -/-/-/+ -/-/+ -/+ -/-/-/+ -/-/- +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ +/+ -/+ -/+ +/+ -/-/-/+/+ -/-/-/-/+/+ -/+ +/+ -/+ -/+ -/-/-/+ -/-/+ -/-/+ -/- +/+ -/+/+ +/+ -/+ -/+ -/+ -/+/+ -/+ -/-/+ -/+ -/-/-/-/-/-/+ -/+ -/+ * */* * * Ricchetti et al.2004, cited * * * * * NUMTs as genetic markers NUMTS as genetic tools to follow the geographic distribution of species or populations and to resolve phylogenetic ambiguities Podnar et al 2007 J Mol Evol 64: 308-320. Unusual origin of a nulcear pseudogene in the Italian wallizard: intergenomic and interspecific transfer of a large section of the mitochondrial genome in the genus Podarcis (Lacertidae). Vartanian JP, Wain-Hobson S. 2002 Proc Natl Acad Sci U S A 99: 7566-7569. Analysis of a library of macaque nuclear mitochondrial sequences confirms macaque origin of divergent sequences from old oral polio vaccine samples. Specie-specific NUMTs Human-specific NUMTs Ricchetti et al.2004 PLoS Biology, 2:96-1313 Hazkani-Covo & Graur 2007 Mol. Biol. Evol. 24: 13-18 PCR amplification and sequence analysis of NUMTs from humans and chimpanzees NUMT A 1 NUMT code 74 90 192 53 132 8919-8992 8447-8542 16386-16552 1762-1814 608-739 2 3 3 4 4 4 4 272 76 1323 93 131 152 240 94 96 95 96 96 94 91 6735-7006 4349-4424 1392-2714 14966-15058 958-1088 2895-3046 2221-2460 712 389 487 326 300 286 868 a 984 465 1810 419 431 438 1108 4 342 94 9323-9664 319 a 661 5 949 79 6591-7902 919 5 8781 88 6388-15168 507 * 5 6 7 7 7 8781 527 100 106 164 88 90 98 94 96 6388-15168 2414-2930 1609-1704 12946-13051 2414-2570 709 338 406 270 751 1236 438 512 434 7-505 8-59 8-84 * * * 7 8 8 505 59 84 85 94 98 1708-2212 803-861 14846-14929 260 638 472 765 697 556 21 8-1470 11-72 11-163 11-2451 12-68 12-89 13-75 13-123 13-256 14-1023 17-69 17-653 18-156 18-192 20-70 22-47 X-267 * * 8 11 11 11 12 12 13 13 13 14 17 17 18 18 20 22 X 1470 72 163 2451 68 89 75 123 256 1023 69 653 156 192 70 47 267 96 98 96 93 94 98 94 91 99 93 97 95 94 97 92 100 95 8405-8538 14645-14716 6636-6798 518-2968 4236-4303 3786-3874 9508-9578 5103-5225 978-1233 5578-6600 10128-10195 6812-7464 14366-14521 7969-8160 12947-13016 6176-6222 684-950 406 316 514 2851 419 567 331 448 602 1265 261 1105 530 396 515 385 560 21 244 351 400 351 478 260 325 346 242 192 452 374 204 445 338 293 Y-66 * Y 66 100 6494-6559 186 Y-71 * Y 71 100 1268-1338 361 * * 2-272 3-76 3-1323 4-93 4-131 4-152 4-240 * * 4-342 * * * * * 5-949 5-8781 up d 5-8781 dw 6-527 7-100 7-106 7-164 d * * * * * * Y-3107 11-541 h 3 size (bp) 1 1 1 2 2 5 mt coord Y 3107 83 1155-4863 11 541 94 16074-60 6 7 PCR size PCR size no NUMT NUMT 344 418 927 1017 825 1017 330 383 365 497 a 8 9 +/+ 2 21 21 4 20 10 11 PCR amplification -/+/sequenced 14 5 1 0 0 1 0 0 2 5 12 2 0 1 1 12 Chimp + + - 21 21 20 21 21 21 21 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 2 0 21 0 0 0 - 0 0 1 + 0 0 3 + 0 0 0 0 0 0 0 0 0 0 2 0 1 0 1 + + + 0 0 0 0 0 0 0 0 0 + + - 21 21 21 21 1 19 21 21 0 21 21 21 3 21 21 21 0 0 0 0 0 13 0 0 0 21 0 0 0 12 0 0 0 0 0 0 0 0 7 2 0 0 0 0 0 0 6 0 0 0 2 0 0 0 0 0 0 0 0 0 0 0 1 5 0 2 0 + + + - 252 0 ND 13 f 1 - 432 0 8 g 13 g 2 - f a 21 c b 21 c b 21 c 21 21 21 21 e 21 21 b 195 C B 4 % identity 100 94 89 100 99 1-74 1-90 1-192 2-53 2-132 2 chr c f f 467 0 ND 13 736 4 8 8 + + - 0 + 0 ND Ricchetti et al.2004, cited Human-specific NUMTs in human chromosomes 30 total n° of NUMTs 250 20 200 15 150 10 100 5 50 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 X Y chromosome size (in Mbp) n° of NUMTs human-specific NUMTs 25 No strict correlation between chromosome size and nb of NUMTs Ricchetti et al.2004, cited Distribution of human-specific NUMTs in chromosomes 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 X Y Ricchetti et al.2004, cited Colonisation rate of the human genome 27 NUMTs are specific to humans and have colonised human chromosomes in the last 4-6 Myrs Ricchetti et al 2004, cited 5000 Alu sequences have colonised human chromosomes in the last 4-6 Myrs from: Batzer & Deininger et al, 2002 Nature. Genet. Rev. 3, 370 180 Alu seq : 1 NUMT NUMTs as potential mutagens Insertion sites of NUMTs in the human genome exon 25 intron promoter intergene number of NUMTs 20 15 10 5 0 NUMTs common to humans and chimpanzees human-specific NUMTs NUMTs with insertion polymorphism in humans Ricchetti et al.2004, cited NUMTs preferentially insert in genes (in humans) 80% NUMTs inserted in genes genes (with introns) < 25% of the human genome Diseases associated with the insertion of NUMTs Diseases associated with the insertion of a NUMT include disorders as diverse as: • a sporadic case of the Pallister-Hall syndrome, a multiple congenital anomaly characterised by hypothalamic hamartoma and bone deformities Turner C et al. 2003. Hum Genet 112, 303-9 Diseases associated with the insertion of NUMTs Diseases associated with the insertion of a NUMT include disorders as diverse as: • a familial plasma factor deficiency, a severe type I factor VII deficiency resulting in severe bleeding Borensztajn K et al. 2002. Br J Haematol 117, 168-71 Diseases associated with the insertion of NUMTs Diseases associated with the insertion of a NUMT include disorders as diverse as: • a mucolipidosis Type IV, showing a moderate phenotype of this usually severe neurodegenerative disorder Goldin E et al. 2004. Hum Mutat. 24, 460-5 Diseases associated with the insertion of NUMTs Diseases associated with the insertion of a NUMT include disorders as diverse as: • a familial bipolar affective disorder associated to a constitutional chromosomal translocation with a NUMT at the junction site Willett-Brozick JE et al. 2001 Hum Genet 109, 216-23 Diseases associated with the insertion of NUMTs The diversity of these pathologies reflects a mutagenic process that can target a large variety of genes. Three diseases related to NUMT insertions Disease NUMT severe type I factor (F) VII deficiency (rare bleeding disorder, family history) 251 bp NUMT insertion in IVS acceptor splice site Borensztajn et al, 2001 Brit. J. Haemat. 117, 168 sporadic case of Pallister-Hall syndrome (de novo insertion) 72 bp NUMT insertion into exon 14 of the GLI3 gene Turner et al, 2003 Hum. Genet. 112, 303 bipolar affective disorder (cosegregation in family pedigree) Willet-Brozick et al, 2001 Hum. Genet 109, 216 41 bp NUMT insertion at the breakpoint junction of a reciprocal constitutional translocation t(9;11)(p24;q23) Disease related to NUMT insertion: sporadic case of Pallister-Hall syndrome (GLI3) ctgcccagcctgctcagcctcacgcccgcccagcagtaccgcctcaaggccaag wt ctgcccagcctgctcagcctcacgcccggtctaacaacatggctttctcaactttt aaaggataacagctatccattggtcttaggccccaaaaattttcccagcagtaccg +NUMT cctcaaggccaag LPSLLSLTPAQQYRLKAK LPSLLSLTPVSNNMAFSTFKG* from: Turner et al, 2003 Hum. Genet. 112, 303 wt +NUMT Impact of new NUMT and Alu insertions on human diseases 17 known disease-related Alu insertions 1,500,000 Alu seq/ human genome 4 known disease-related NUMT insertions 211 NUMTs /human genome NUMT insertions can modify the exon/intron pattern Some NUMT insertions in genes hypothetical protein a 1 2 b 1 2 3 4 3 NUMT 12-89 hypothetical protein a 1 2 3 4 5 b 1 2 3 4 5 6 NUMT 17-653 hypothetical protein a 1 2 3 4 5 6 7 8 b 1 2 3 4 5 6 7 8 10 9 NUMT 5-8781 protein Q8N7L5 a 1 2 3 4 5 6 b 1 2 3 4 5 6 Ricchetti et al.2004, cited 7 7 8 NUMT 1-74 Generation of novel nuclear exons by NUMTs Nuclear insertions of organelle DNA in yeast, H. sapiens, Arabidopsis, and rice: 45 insertions contributed sequences to a total of 49 protein-coding exons in 34 genes. Noutsos et al. 2007 Trends Genet. 23:597–601 NUMTs as regulatory sequences? Discussion on unpublished data by Chatre and Ricchetti, 2009 5. How do nucleic acids move to the nucleus? • lysis of mitochondria ? • capture of cytoplasmic nucleic acids ? • illegittimate transport ? • membrane fusion ? Mitochondria-nuclear interactions Human HeLa cells. Subpopulation of perinuclear mitochondria under normal growth conditions. By Laurent Chatre, Institut Pasteur Perspectives Analysis of the possible regulatory function of NUMTs in other eukaryotes, including humans Analysis of mt-nuclear interactions and mt DNA release under normal growth and stress conditions Acknowledgements Laurent Chatre Benjamin Montagne Cecile Fairhead Fredj Tekaia Bernard Dujon Unité de Génétique Moléculaire des Levures Qu i ck Ti me ™a nd a TIF F (Un co mpre ss ed )d ec omp res so r a re ne ed ed to s ee th i s pi c tu re. QuickT ime ™an d a TIFF ( Uncomp res sed) deco mpre ssor ar e need ed to see this pictur e.