Mécanismes moléculaires d'induction et d'action antivirale
advertisement

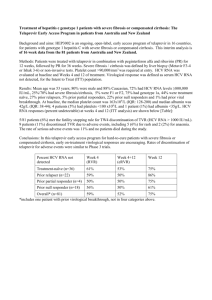
Interaction of HCV with Interferon and the innate immune response Eliane F.Meurs Pasteur Institute, Paris INTERFERONS Interferons belong to the family of type II cytokines ( 27 cytokines, 12 receptors). Interferons are proteins naturally induced by the organism in response to a situation of stress, such as infection with bacterial or viral pathogens. IFNs are involved in the resistance of cells to a viral infection,in growth regulation and activation of immune response. Introduction INTERFERONS Identified through their antiviral properties (Isaacs and Lindenman, 1957) Influenza Virus Chicken chorio-allantoïc Membranes Culture medium Heating, Filtration reduced viral production Production of virus Culture medium contains some protecting substance: Interferon Cloned in 1980 Interferon messenger RNA: translation in heterologous cells. de Maeyer-Guignard J, de Maeyer E, Montagnier L. A viral inhibitor with the characteristics of mouse interferon is produced by avian and simian cells preincubated with RNA extracted from interferon-producing mouse cells. Similarly, RNA extracted from interferon-producing monkey cells induces a monkey interferon-like substance in avian cells and also in a line of simian cells, VERO, which normally lacks the capacity toproduce its own interferon. In both cases, the RNA effect is inhibited by treatment of the receptor cells by cycloheximide,but not by actinomycin D. We conclude that interferon messenger RNA has been translated in the receptor cells. Thus, the production of interferon in heterologous cells can be used as a sensitive assay of interferon messenger RNA. Proc Natl Acad Sci U S A. 1972 May;69(5):1203-7 IFN-producing Monkey cells Extraction RNA IFN mRNA translated in the heterologous VERO cells Incubation RNA with VERO cells (cells which cannot make own IFN) Resistance to viral infection Inhibited by cycloheximide Not by actinomycin The type I IFN system Bacteria, virus IFNAR Activation JAK/STAT Activation IRF3 Activation NF-B Induction IRF7 IFNAR Activation JAK/STAT Activation IRF3/IRF7 IFN- subtypes IFN some early ISGs Induction ISGs Induction IFN (Several subtypes) Amplification phase Innate immune response antiviral and Antiproliferative state Activation of adaptive immune response: -Maturation of DCs (induction CD80,CD86, CD40) -Stimulation NK -Stimulation of CTL differentiation Interferons and their use in therapeutics The different IFN types Type I IFNs 9p21p22p23 IFN IFN IFN IFN 1 gene, 35% identity with 13 gènes, 5 pseudogenes,70 % identity 1 gene, 5 pseudogenes, 55% id. , 29% id. 1 gene IFN 1 gene, 30% homol with , or (6Mb away from IFN cluster= separated evolution) IFN (limitin) 1 gene, 30% id.avec IFN , and (mice) IFN 10 genes; 55% identity with , 70% with (sheep, cattle) Type III IFNs 19q13,13 1 (IL-29), 2 (IL-28A), 3 (Il-28B) homologies with IFNs and with IL-10 (Only 2 and 3 in mice) IFN Type II IFNs IFN 12q24,1 1 gene; No identity with other IFNs Human IFN- genes and proteins 12 distinct IFN-s from 14 genes IFNA1 IFN-D, IFN-1 IFNA2 IFN-A (IFN-2a), IFN-2 (IFN-2b), IFN-2c IFN-4a (IFN-76), IFN-4b IFNA4 IFNA5 IFNA6 IFN-G, IFN-5, IFN-61 IFN-K, IFN-6, IFN-54 IFNA7 IFN-J, IFN-J1, IFN-7 IFNA8 IFNA10 IFNA13 IFN-B2, IFN-B, IFN-8 IFN-C, IFN-10, IFN-1, IFN-6L IFN-12 ( sequence identical to IFN-1) IFNA14 IFNA16 IFN-H, IFN-H1, IFN-14 IFN-WA, IFN-16, IFN-O IFNA17 IFNA21 IFNAP22 IFN-I, IFN-17, IFN-88 IFN-F, IFN-21 IFN-E IFN-2: predominantly used in clinic Petska, S review JBC,2007 Therapeutic use of Interferons IFN-s : Hairy cell leukemia, malignant melanoma, follicular lymphoma, genital warts, AIDS-related Kaposi’s sarcoma,laryngeal papillomatosis, chronic hepatitis C,chronic hepatitis B Interferon Alfa-2a (Roferon-A; Hoffnung); Interferon Alfa-2b (Intron-A; Schering) pegylated Alfa-2a (Pegasys; Roche); Pegylated Alfa-2b (Peg-Intron; Schering) Interferon Alfa-2b+ Ribavirin (Rebetron); Interferon alfacon-1 (Infergen) Interferon Alfa-2b fused to Albumin (Albinferon) IFN-: Multiple sclerosis, Cervical intra-epithelial Neoplasia Interferon beta-1b (Betaseron); Interferon beta-1a (Avonex); Soluferon (preclinical testing;VPM) IFN-: In Phase I studies Peg-Interferon (Zymogenetics) IFN- Chronic granulomatous disease, severe malignant osteopetrosis Interferon gamma-1b (Actimmune) Towards new IFN technologies -Generate new IFN molecules using gene shuffling technology: AV Th1 Binding Antiprolif. Brideau-Andersen et al, PNAS, 2007 Maxyalpha in clinical trials (Maxigen) Interferon treatment and HCV Monotherapy (Interferon, PegInterferon) Bitherapy (PegInterferon + Ribavirin) Adding specific antiviral compounds 20 to 50% SVR 50 to 80 % SVR can we cure? Ex: Antiviral effect of Telaprevir (VX-950), an anti-HCV NS3/4A protease inhibitor ; a 15-day clinical trial Placebo+ PegIFN Telaprevir Telaprevir + PegIFN Forestier, N., Hepatology,2007 Mechanism(s) of IFN Induction: IFN INDUCTION PAMPs (MAMPs) Bacteria, virus Pathogen-Associated Molecular Patterns Microorganism-Associated Molecular Patterns cell PRRs Pattern-recognition receptors IFN PAMPs + PRRs IFN Adapter proteins Activation of kinases Activation of transcription factors Gene induction PAMPs Bacteria Fungi LPS PhospholipoLipoproteins mannan Peptidoglycans GlucuronoxyloGlycolipids mannan Flagellins DNA Unmethylated CpG Staph aureus Trepon.maltophil. listeria Candida albicans Aspergillus fumig. S.cerevisiae Pneumocustis car. Protozoa Glycosylphosphatidyl Inositol Glycoinositolphospholipids Trypanosoma cruzi T brucei Toxoplasma gondii Leishmania major Plasmodium falcip. Viruses Envelopes ssRNA dsRNA ssDNA DNA Influenza Measles VSV Rabies virus HCMV HSV-1 RSV MMTV WNV HCV Toll-like receptors or TLRs Peptidoglycan (G+) Lipoproteins (G-) Peptidoglycan(G+) TLR1 LPS(G-) RSV-F Flagellins (G+) TLR4 TLR2 TLR5 Lipoproteins (G+) mycoplasms TLR6 TLR10 Not present in mice Plasma membrane Endosomes dsRNA TLR3 Loxoribine Imiquimod ssRNA TLR7 Imiquidiazoline Unmethylated CpG ARNsb viral Resiquimod ssRNA TLR8 TLR9 Not functionnal in mice Extracellular domains of TLRs : 19-25 tandem copies of LRR A common intracellular domain : TIR, 200 residus, assembly platform All TLRs use Myd88 as adapter, exceptTLR3 Signaling pathways activated by TLR 3,4,7,8,9 Expression pattern of TLRs The 10 human TLRs are expressed in all tissues with various levels Highest levels TLR1 TLR2 TLR3 TLR4 TLR5 TLR6 TLR7 TLR8 TLR9 TLR10 kidney, lung, spleen lung, spleen Placenta, and ubiquitous spleen and ubiquitous ubiquitous ubiquitous lung,placenta, spinal cord, spleen lung, spleen skeletal muscle, spleen spleen, thymus Cytosolic PRRs and PRMs the NOD-Like Receptors or NLR the RIG-I-Like Receptors or RLR the DNA sensors the NOD-Like Receptors or NLR NALP7 NALP14 NALP5 NALP12 NALP3 Bacteria, MAMPs,DAMPs NOD1,2 NALP4 NALP9 NALP11 NALP13 NALP.. NOD9/NLRX1 NF-kB, MAPK Inflammosome ROS IL1 NALP8 NALP1 NALP6 NOD3 NOD9/NLRX1 Antimicrobial defence NOD2 NOD1 NOD27 CIITA IPAF NAIP Tattoli, I et al, Embo reports 2008 NALP2 the NLR Contain LRR and nucleotide binding domain (NBD) Cytosolic PRRs: the RIG-I-Like Receptors or RLR Treatment with IFN Virus Human K562 cells Human K562 cells (deletion locus IFN type I) Generation cDNA library activation IRF3 No IRF3 activation Poly(I)-poly(C) Murine L929 cells Induction IRF-luc reporter Out of 100 000 cDNA, isolation of one clone encoding for a DExD/RNA helicase: RIG-I Yoneyama et al, 2004 Kato,H et al,July 2005,Immunity The DExD/H RNA helicase RIG-I/Mda-5/LGP2 family CARD RNA helicase 925 RIG-I DEAD motif: in the ATP-binding domain 23% identity with RIG 35% identity with RIG MDA5 LGP2 1025 678 31% identity with helicase domain of RIG 41% identity with helicase domain of MDA5 No CARD domain These RLR are inducible by IFN Yoneyama et al, J Immunol, 2005 Viruses and dsRNA Induce IFN through the RLRs: Sensors of non-self RNA In vitro transcribed RNAs Infection by NDV, VSV, SeV, Rabies,Influenza Measles leader RNA HCV 5’ and 3 UTR Poly(I)-poly (C ) Infection by picoRNAvirus (EMCV, Theiler, Mengo) activation RIG-I Importance of a triphosphate at the 5’ minimun size 21 bp activation MDA5 Poly(I)-poly (C ) dsRNA Other RNAs? Binding LGP2 Differential regulation of RIG-I and MDA5? LGP2-/- mice: susceptible to EMCV Less or highly susceptible to VSV? Kato, H. Nature 2006 Hornung, V et al, Science,2006 Pichlmaier,A.Science, 2006 Plumet, S. PlosOne, 2007 Venkataraman,T, J Immunol,2007 Kato, Oxford meeting, sept 2007 Detection of structurally distinct RNA species by RIG-I and triggering of signalling RNA binds to RIG-I C terminal domain CTD; NMR studies : binding of RNA into a basic cleft RIG-I CTD helicase dsRNA 5’pppssRNA CARD helicase ATP Signalling CTD CARD + polyIpolyC CTD helicase CARD Abortive T.Fujita, Oxford ISICR meeting, sept 07 The HCV RNA act as a PAMP to activate RIG HCV RNA hel. RIG-I 3' CARD CTD 5’ PPP 5’ PPP IFN X Poly(U/UC) replicatio n T55I RIG-I Activation dependent on U/UC region at its 3’end and of 5’ppp end CARD RNA helicase 925 The huh7-5 cells have an endogenous defective RIG-I which explains, at least in part, their permissivity to HCV expression Sumpter,R.J Virol, 2005; Saitoh and Gale,M;Nature 2008 MDA5 binds to long and RIG-I to short dsRNA structures Atomic Force Microscopy Kato,H et al, JEM 2008 ATPase MDA5 ATPase RIG-I long dsRNA + - short dsRNA + Activation of RIG-I or MDA5 by RNA VIruses Genome VSV Replicative Intermediates EMCV ss RNA 8 kB 10 segments dsRNA 3.9 kB 2.2 kB 1.2 kB Kato,H et al, JEM 2008 + + ds RNA 8 kB ss RNA, 5’ppp HCV ss RNA, 5’ppp Reo MDA5 ss RNA 11 kB DI dsRNA 2.2 kB Flu RIG-I + 5’ppp + poly (U/UC) + + + RIG-I’s mediated antiviral activity requires its ubiquitination through TRIM25 Gack et al, Nature, 2007 Effect of deficiency of RIG-I, MDA5 and LGP2 on viral infections RIG-I KO: mostly embryonic lethal at 12.5 to 14 days, few mice born alive, die within 3 weeks. Experiments with isolated cells : RIG-I essential for induction of IFN by RNA viruses in fibroblasts and DCs Experiments with RIG-I -/- mice: highly susceptible to infection with JEV, resistant to infection with EMCV Kato, H. et al, Immunity, 2005 Kato, H. et al, Nature, 2006 MDA5 KO: mice alive and healthy Experiments with MDA5 -/- mice: highly susceptible to infection with EMCV, resistant to JEV Kato, H. et al, Nature, 2006 LGP2 KO: viable Experiments with MEFs: in response to polyIpolyC, the loss of LGP2 increases IFN production Experiments with LGP2-/- mice: resistant to VSV infection (in accord with negative regulation of RIGI) sensitive to EMCV infection ( why?) Venkataraman, T. J Immunol, 2007 Viral infection dsRNA DNA TLR3 TLR viral RNA RIG-I/MDA5 TRIF MAVS Facteurs transcription AP-1 IRF3 CBP/p300 NF-B IFN- Bacterial components TLR IPS-1/MAVS/VISA/CARDIF The adapter between RIG-I/MDA5 and the downstream IFN-inducing kinases 1st identification in 2003: a gene with no known precise function Large-scale identification and characterization of human genes that activate NF-B and MAPK signaling Pathways Matsuda,A et al.( 2003) Oncogene 22 3307-3318 Identified in 2005 as RIG-I adapter by S.Akira (Kawai,T et al, Nature Immunol,29 Aug.2005 ): High throughput screening IPS-1 ( IFN- Promoter Stimulator-1) Z.Chen (Seth,R. et al, Cell, 122,1-14, Aug.2005) Blast search with domain CARD-like of RIG-I MAVS (Mitochondrial AntiViral Signaling) H.B.Shu (Xu,L-G et al, Mol Cell, 19 1-14, Aug. 2005) Cloning and analysis of the NF-B-inducing ability of the Matsuda gene VISA (Virus-Induced Signaling Adapter) J.Tschopp (Meylan,E. et al, Nature, sept 2005) Blast search with domain CARD-like of RIG-I CARDIF (CARD adapter Inducing IFN-) The RIG-I/MDA5-mediated IFN induction pathway Virus 5’pppRNA, small dsRNA (RIG-I) long dsRNA (MDA5) cytosol CARD IPS/VISA/MAVS/CARDIF CARD TM CARD PRO TRAF6 TAB/TAK IKK MAPKs P P IKK IKK ? Ub Ub Ub Ub AP1 IB Mitochondria TRAF3 TANK TBK1 IKK P P IRF-3 P50/p65 NF-B P AV state inflammation IFN- P AP1P P NF-B NF-B IRF-3 Nucleus Pro-inflammatory cytokines The IRF3/IRF7-phosphorylating kinases TBK1 and IKK 19 299 305 383 730 Kinase domain ULD Coiled-Coil Coiled-Coil 19 299 305 383 717 Kinase domain ULD Coiled-Coil Coiled-Coil TBK1 IKK Homologous, yet with some differences -Deletion of TBK1 but not of IKK gene is lethal (Bonnard, M et al, EMBO, 2000; Hemmi, H et al, J Exp Med, 2004 ) -TBK1 more important role than IKK in IFN induction in response to different stimuli. Yet, double KO cells have complete abolition of IFN induction (Hemmi, H et al, J Exp Med, 2004 ) -Expression of TBK1 is constitutive. IKK is inducible in response to IL-1, PMA and virus through NF-B and c/EBP (Shimada, T et al, Int.Immunol,1999) -IKK induces a subset of IFN-induced genes (ADAR-1) through ser708 phosphorylation of STAT1 (tenOever,B. et al, Science, 2007) IRF3 Ubiquitous expression; Localization: inactive in the cytosol, active in the nucleus TLR3 TLR4 RIG-I/MDA5 IRF3 (Qin et al; Takahasi et al, Nature Struc Biol;2003) TBK1 and IKK are the IRF3-phosphorylating kinases Activation TBK1/IKK Phosphorylation IRF3 1 427 382 414 SRR (serine rich region) 7 phosphate acceptor sites -GGAS385S386LENTVDLHIS396NS398HPLS402LT404S405DQYKAYLQD- essentiels (S A abolishes activity) non essentiels individually (S A : normal phenotype) Induction IFN inhibited when all 5 residues are mutated Phosphorylation of IRF3 liberates its DNA binding activity Sharma, S et al, Science 2003; Fitzgerald, K; Nature Immunol, 2003; Higgs, R and Jefferies,C. 2008 The IRF family IRF-1 IRF-2 DBD (DNA Binding Domain) P IAD (IRF association Domain) P Homologies with C-ter of SMADs C-ter auto-inhibition domain Proline-rich sequence IRF-5 IRF-6 P Phosphorylation site IRF-8 (ICSBP) IRF-9 (ISGF3g/p48) IRFs have distinct roles in the development and function of immune cells IRF-4 (PIP) IRF-3 IRF-7 P P Honda, K and Taniguchi, T, review,2006 Viral infection dsRNA DNA TLR3 TLR viral RNA RIG-I/MDA5 TRIF MAVS Facteurs transcription AP-1 IRF3 CBP/p300 NF-B IFN- Bacterial components TLR OVERALL STRUCTURE OF THE IFN ENHANCEOSOME PRDIV PRDI PRDIII PRDII Panne, D et al, Cell,2007 General scheme of IFN induction by viruses and dsRNA virus dsRNA Viral RNA RIG-I/Mda5 TLR3 TRIF MAVS p38MAPK/JNK AP-1 IKK NF-B TBK1/IKK IRF-3 IFN CXCl-10 Early ISGs ISG15,etc… JAK/STAT IRF7 (reinduced by IFNs) All IFN-s + ISGs Including RIG-I/MDA5/LGP2, TLR3 IFNAR Induction de gènes par IRF3: consensus AANNGAAA IRF3 + CBP/p300 c-Jun/ATF2 NF-B IFN GAGAAGTGAAAGTG -104 PRDI-III PRDIV PRDII -55 ISG56 IFN CAAAGTGAAAGG GGAAAGCCAAAGG -92 -117 ISG15 IFN CGGGAAAGGG GAAACCGAAACTGA -165 -143 IFN1 IRF7 ATGAAGGAAAGCAAAAACAGAAATGGAAAGT Cell-mediated positive and negative regulation of the IFN induction pathway Moore, C. and Ting,J. Minireview, Immunity 2008 Gack et al, Nature, 2007 IFN inducing pathways: targets for viral inhibitors viral RNA MDA-5 TRIF RIG-I MAVS AP-1 NF-B Capside 3 E3L NSP3 Reovirus Vaccinia Rotavirus V protein NS1 Paramyvovirus Influenza NS3/4A HCV IRF3 vIRF1, vIRF3 HHV8 E1A Adenovirus (binds CBP/p300) CBP/p300 IFN- E6 (binds IRF3) NSs (inhibits TFIIH) Papillomavirus Rift valley fever Bunyavirus The HCV RNA act as a PAMP to activate RIG… …yet HCV inhibits the IFN inducing pathway Activation of IFN Signaling pathway NS3/4A Inhibition phosphorylation IRF3 and of its migration to nucleus Inhibition of IFN signaling pathway Foy et al, 2003 Helicase domain C NS4A cofactor Catalytic domain protease domain How does HCV NS3/4A interferes with the IFN inducing pathway? NS3/4A cleavage site: P6-P5-P4-P3-P2-P1.....P’1-P’2-P’3-P’4 Consensus sequence: D- X- X- X- X- C.....S- X- X- L (E) (T) (A) (W/Y) N from De Francesco R, 2003 Sequence of the NS3/4A-mediated cleavages in HCV polyprotein from different genotypes Name H77 HCV-N JFH1 NZL1 ED43 SA13 6a33 Genotype (GI ) NS3 / 4A 1a (GI:2316097 ) MSADLEVVT STWVLVGG 1b (GI:23957856) MSADLEVVT STWVLVGG 2a (GI:13122261) MQADLEVMT STWVLAGG 3a (GI:514395) MSADLEVTT STWVLLGG 4a (GI:2252489) MSADLEVVT STWVLVGG 5a (GI:3660725) MSADLEVIT STWVLVGG 6a (GI:57791993) MSADLEVIT STWVLVGG Meurs& Breiman, WJG,2007 NS4A/ 4B QEFDEMEEC SQHLPYIE REFDEMEEC ASHLPYIE EAFDEMEEC ASRAALIE QQYDEMEEC SQAAPYIE QQFDEMEEC SKHLPLVE QQFDEMEEC SASLPYMD QQFDEMEEC SRHIPYLAE NS4B/ 5A ISSECTTPC SGSWLRDI INEDCSTPC SGSWLRDV ITEDCPIPC SGSWLRDV INEDYPSPC SDDWLRTI INEDCSTPC STPCAESW IGEDYSTPC DGTWLRAI VNEDTATPC ATSWLRDV NS5A/ 5B reference ADTEDVVCC SSYSWTGA Kolykhalov et al, 1997 EAGESVVCC SMSYTWTG Beard et al, 1999 EEDDTTVCC SMSYSWTG Kato et al, 2001 SEEQSVVCC SMSYSWTG Sakamoto et al, 1994 SGSEDVVCC SMSYSWTGChamberlain et al, 1997 SDEDSVVCC SMSYSWTGBukh et al, 1998 SDQDDVVCC SMSYSWTGZhou et al, 2004 Is it through its protease activity? HCV NS3/4A cleaves IPS-1/VISA/MAVS/CARDIF 1 TM MAVS CARD PRO 540 508 EREVPCHRPS Meylan et al (2005) Nature NS3/4A cleaves also TRIF dsRNA TLR3 TRIF 372 N C 192 DGVSDWSQGCSLRSTGSPAS 326 PILEPVKNPCSVKDQTPLQL 372 PPPPPSSTPCSAHLTPSSLE 1 TM MAVS Li, K et al (2005) PNAS CARD PRO 540 508 EREVPCHRPS Meylan et al (2005) Nature -MAVS is delocalized from the mitochondria to the cytosol in cells transfected with NS3/4A, infected with HCVcc and in liver biopsies from chronic hepatitis patients (Lin et al, 2006; Loo et al, 2006) -The BILN 2061 NS3/4A inhibitor can restore the MAVS-mediated IFN induction in cells Infected with HCVcc (Cheng et al,2006) MAVS Cont MAVS + NS3/4A Lin et al, 2006 Cheng et al,2006 Distinct localizations of IKK and TBK1: IKK associates with the mitochondria IKK MITO Merge TBK1 MITO Merge IKK IKK MAVS MAVS Colocalization of Cardif and IKK IKK MAVS Merge IKK MAVS Merge Merge Merge Disruption of the IKK/ Cardif complex localization by expression of NS3/4A +NS3/4A Lin et al, J Virol 2006 The RIG-Ipathway in liver biopsies from HCV-infected patients 17 patients with chronic HCV: -9 under no treatment -8 non responders to treatment 12 controls (other pathologies) Patient number 1 2 3 4 5 6 7 8 9 ALT (IU/L) 78 31 75 93 24 23 70 37 69 10 11 12 13 14 15 16 17 137 42 27 47 47 70 29 36 AST (IU/L) 29 18 47 54 17 16 34 27 28 Viral load HCV (IU/mL) genotype 5,00E+06 1b 3,45E+05 1b 2,36E+05 4d 2,02E+05 1 2,00E+06 1b 2,00E+05 1a 1,00E+06 4 ND 5,46E+07 1a 76 26 23 32 36 41 27 25 1,12E+05 2,00E+08 2,00E+07 1,20E+07 2,40E+04 5,91E+05 9,36E+06 4,00E+05 dsRNA-activated RNA Helicases Mitochondrial adapter Genes analysed by qRT-PCR Genes used for normalisation: TRIM44,HMBS, BC002942 IRF3-phosphorylating kinases IKK -induced genes 4 1b 1b 1b 1b 1b 1a 1b Knodell Index 1+1+1+0 1+1+1+0 3+0+3+0 1+3+4+0 0+1+1+0 NA 1+1+1+0 1+1+3+1 1+1+1+0 Treatment IFN/Ribavirin No No No No No No No No No Time after treatment 3+4+3+3 NA 1+1+3+1 1+1+3+1 1+1+3+0 3+3+3+1 1+1+3+0 1+1+3+1 Yes Yes Yes Yes Yes Yes Yes Yes 2 years 6 years 9 years 5 months 4 years 7 years 5 years 11 years RIG-I, MDA5, LGP2 MAVS IKK, TBK1 IFN, ISG15, CCL3 Analysis of liver biopsies: results 1 NS NS 9 8 7 6 5 4 3 2 1 Cont HCV HCV.NR IFN- TBK-1 NS NS NS 6 NS 5 4 3 2 1 Cont HCV HCV.NR Relative RNA Expression NS Relative RNA Expression x 10 -2 Relative RNA Expression x 10 3 MAVS 8 7 6 NS NS 5 4 3 2 1 Cont HCV RNA expression levels of MAVS, IFN- and TBK-1 are not significantly affected in HCV-infected patients HCV.NR Analysis of liver biopsies: results 2 9 8 7 6 5 4 3 2 1 MDA5 P=0.01 P=0.015 NS Cont HCV 25 P<0.001 P=0.06 P=0.01 20 15 10 5 HCV.NR Cont P=0.005 NS NS 7 6 5 4 3 2 1 Cont HCV 12 P=0.016 NS NS 10 8 6 4 2 Cont HCV HCV.NR ISG15 HCV.NR Relative RNA Expression x 10 -1 Relative RNA Expression LGP2 HCV HCV.NR IKK Relative RNA Expression x 10 -1 RIG-I Relative RNA Expression Relative RNA Expression x10 1 HCV interferes in vivo with the dsRNA-RIG-I/ IKK pathway P<0.001 P<0.001 NS 8 7 6 5 4 3 2 1 The RNA expression levels of RIG-I, MDA5, LGP2, IKK and ISG15 are downregulated in HCV-infected patients. Cont HCV HCV.NR Vilasco et al, Hepatology, 2006 HCV can both provoke and inhibit the IFNinducing pathways Treatment of patients with exogenous IFN HCV Activation IRF3 Activation NF-B IFNAR Activation JAK/STAT Induction IRF7 IFNAR Activation JAK/STAT Activation IRF3/IRF7 IFN- subtypes IFN some early ISGs Induction ISGs Induction IFN (Several subtypes) Amplification phase antiviral and Antiproliferative state Can HCV affect the response to IFN and some antiviral mechanisms? Response of cells to Interferons THE JAK/STAT SIGNALING PATHWAY ligand Receptor Signal Transducers and Stat Activaters of Transcription Tyr P Jak 4 JAKs 7 STATs STAT1 STAT2 STAT3 STAT4 STAT5A STAT5B STAT6 Janus Activated tyrosine Kinase ISG TYK2 JAK1 JAK2 JAk3 THE IFN RECEPTOR R1 CHAIN : - responsible for signal specificity -long intracellular domain, associates to JAKs -is phosphorylated on tyrosyl residue -recruits STAT through their SH2 domains R2 chain : -only used for signal transduction -short intracellular domain -Recruits JAK -Allows JAK cross-activation IFN R1 R2 Tyr P IFN- fibronectin type III Il-20 FVIIa TF IFN- Il-10 IL-10R2 Il-22 IL-10R2 IFNAR2 IFNAR1 IFN-R1 IL-10R1 IFN-R2 IFN- R1 IL-10R1 IL-10R2 Il-26 Il-24 IL-10R2 IL-10R2 IL-20R2 IL-20R1 IL-22R1 Il-19 IL-20 IL-24 IL-20R2 IL-20R1 IL-22R1 The cytokine class II receptor superfamily Kotenko, S and Langer,J. 2004 The different steps of JAK/STAT activation 1 2 IFNAR1 IFNAR1 IFNAR2 Tyk2 Tyr466 y P Jak1 IFNAR2 Tyk2 Tyk2/Jak1 Y Stat2 Jak1 Tyk2/Jak1 y P P Stat2 Tyk2 Jak1 3 4 Tyk2 Jak1 Tyk2/Jak1 yP P Stat1 5 yP P Tyk2/Jak1 P y P Stat2 Jak/Stat activation by IFN-/ IFNAR1 IFNAR2 Tyk2 Jak1 Tyk2/Jak1 P P Stat2 P P P Stat1 Type I IFN activate other Stat complexes IRF9 P nucleus Stat 1:1 Stat 2:1 Stat 1:3 Stat 5:5 Stat 3:3 Stat 2:6 ( in mature B cells) Stat 4:4 (CD4+ Tcells) P ISGF3 P ISRE IFN-Stimulated Genes GGAAANNAAACT …but those bind GAS or GAS-like elements Brierly,M and Fish,E. 2005 Jak/Stat1 activation by IFN- IFN- IFNR2 R1 R1 IFN-R2 Jak2/Jak1 P Stat1 P Type II IFN activate other Stat complexes P P Stat 3:3 Stat 3:1 P P nucleus P P GAS IFN- induced genes TTNCNNNAA The Janus kinases (JAK) family TYK2 JAK1 JAK2 JAk3 -JH: JAK homology domains. Only JH1 has catalytic activity. JH2 has a pseudokinase domain. JH5 to half of JH4: SH2 domain; JH1-3 and half of JH4: FERM domain (4.1, ezrin, radixin, moesin): allows stable association with membrane proximal receptors motifs. Jak1 associates with IFNAR2 and IFNGR1; JAK2 with IFNGR2; Tyk with IFNAR1 -Expression in most tissues, except JAK3 (leukocyte) Jak1 KO mice: die perinatally; tissues defective in response to IL-2, IL-6, IFN and IL-10 Jak2 KO mice: Embryonic lethality (E12.5). Crucial role in erythropoiesis Jak3 KO mice: severe Combined Immunodeficiency (SCID)-like defects due to physical link of Jak3 to C, associated to IL-2 family of cytokine receptors Tyk2 KO mice: modest cytokine defects, type 2 T-cell response. Tyk2 deficient humans: severe allergic phenotype: impaired antimicrobial defense The STAT family STAT1, STAT2, STAT3, STAT4, STATA STATB, STAT6 STAT1 1 N ter 135 Coiled-coil 317 DBD 488 linker 576 SH2 683 TAD 750 Y SSS Contact with DNA Interaction with Dimer-dimer regulatory proteins: interaction IRF9,c-jun, Nmi-1,… and stabilisation P P Y701-P: STAT dimerization nuclear translocation DNA binding Recruitment to YPhosphorylated Receptors And other P-STATs SH2 linker Stability, cycling between cytoplasm and nucleus Favor the formation of STAT1/STAT2 heterodimers and ISGF3 formation DNA binding coiled-coil S708-P , S727-P, S744-P, : Maximal activation Recruitment coactivators: CBP/p300, MCM5, BRCA1 Physiological importance of STAT1 STAT1-knockout mice: high susceptibility to microbial and viral infections and tumor formation STAT1- knockout humans: susceptibility microbial and to viral diseases; death in infancy Kindred B Kindred A Wt/m m/m STAT1wt 1 N ter Wt/m Wt/m 135 BCG HSV-1 Coiled-coil 317 Wt/m m/m DBD BCG virus? linker SH2 488 576 750 603 INFANT 1 1757-1758delAG TAD 683 L600P L706SP INFANT 2 Non conservative substitution Dupuis et al, Nat Gen.2003 Reich et al. Nature Reviews Immunology 2006) Schindler,C and Plumlee,C.Sem. In Cell &dvlpmental Biology 2008 in press conserved Cys WSXWS motif Ig-like domain fibronectin type I D200 class II class I GPI c EPO-R PRL-R IL-9R IL-4R IL-2R IL-5R IL-3R GM-CSFR GH-R IL-7R CNTF-R IL-6R IL-11R gp130 LIF-R KH97 The cytokine receptor superfamily Class I receptors also use the JAK/STAT pathway IFNAR1 IFNAR2 IFN-R1 IFN-R2 IL-10R1 IL-10R2 TF etc.... IL-2 IL-10 EPO IL-4 IL-6 TPO IL-7 IL-11 IL-3 IL-9 OSM IL-5 IL-15 LIF IFN- Leptin IL-21 CNTF G-CSF GM-CSF IL-22 IFN-/ IL-12 IL-23 Prolactin G-H The majority of the cytokines receptors use different JAK combinations leading to activation of multiple STATs Murray, P. J Immunol Review, 2007 IFN IFN JAK1/Tyk2 STAT1:2/IRF9 JAK1/JAK2 STAT1:3 STAT2 STAT1:1 STAT1 Combination of IFN and IFN may enhance AV and immune responses STAT3 IFN may attenuate IFN activation of Stat Huh7 cells + IFN with or without IFN Microarray analysis Ag presentation (20%) Immune-related (11%) Other unknown (35%) Complement activation (6%) Immune cell recruitment (4%) Direct antiviral (3%) Cell growth/apoptosis (12%) Transcription (9%) Blatt, L et al, J IFN and cytok.Res.2005 Radeava,S. Biochem J..2004 Regulators of the JAK/STAT pathway PTP PIAS SOCS ligand SOCS: CD45, PTP, SHP: receptor Suppressor of Cytokine Signaling Jak STAT P Protein tyrosine phosphatases STAT P P SOCS PTP1B, TC-PTP (cytop.), SHP1, SHP2 STAT4 SH2-containing Tyr Phosphatase TC-PTP (nuclear), PIAS1, PIAS3, PIASx, PIASy HDAC STAT1 STAT3 PIAS: STAT4 Protein Inhibitor of Activated STATs SOCS The family of Suppressor of Cytokines Signalling proteins -induced by cytokines; also induced by LPS, isoproterenol, statins, cAMP - act in a negative feedback loop to inhibit cytokine signal transduction -regulate immune responses and maintain immunological homeostasis -8 members: SOCS 1- SOCS 7 and CIS Structure Acts as pseudosubstrate Inhibits the JAK’s Tyr kinase activity (KIR only in SOCS1 and SOCS3) SOCS1 Recruitment of E3 ubiquitin ligase Degradation of proteins Determinant for the target of each SOCS protein Yoshimura,A.Nature Reviews Immunology 2007 Function of SOCS SH2 of SOCS1 bind directly to IFNAR and IFNGR and directly to the activation loop of JAK SH2 of SOCS3 bind directly to gp130-related cytokine receptors No good affinity for activation loop of JAK binds the kinase domain of JAK through Its KIR domain Model: SOCS binds the IFN receptor first and then interacts with JAK Yoshimura,A.Nature Reviews Immunology 2007 Role of SOCS in innate immunity SOCS1-deficient cells and Socs1-/- mice are resistant to viral infections Yoshimura,A.Nature Reviews Immunology 2007 HCV infection Altered expression and activation of STATs and STAT regulators Expression of HCV proteins inhibits IFN induction through Jak/STAT Heim et al, J.Virol, 1999 HCV core protein provoques STAT1 degradation, by interacting with STAT1 SH2 domain Lin,W. Gastroenterol.2005; J Virol, 2006 STAT3 is down-regulated in HCV-infected livers from human patients and in Huh7 cells bearing the full length HCV replicon Larrea, E. et al, Gut. 2005 Non-response to AV therapy is associated with obesity and increased SOCS3 in patients with chronic hepatitis C, viral genotype 1 Walsh,M, Gut, 2005 HCV infection in chimpanzees lead to a type I IFN response. However, deficiency in response to subsequent IFN treatment. Expression of SOCS3 elevated. Huang, Y et al, Gastroenterol. 2007 The IFN-responsive pathway: Target for viral inhibitors Viroceptors (homologs of soluble receptors) IFNAR1 Vaccinia Orthopoxvirus IFNAR2 T antigen (binds Jak1) Tyk2 V protein (blocks Stat1 phospho by Jak1) High induction of SOCS Jak1 Stat1 SOCS P P Stat2 IRF9 P P Core protein (binding to STAT1 SH2 and degradation) nucleus ISGF3 V protein (binding and degradation of STAT) P P ISRE ISGs E7 (binding IRF-9) Murine Polyomavirus Measles HCV, influenza SV5, Mumps (STAT1) Hum.parainfluenza 2 (STAT2) HCV HPV-16 Antiviral action of some IFN-induced genes The IFN-induced genes Virus, dsRNA More than 300 genes induced Activation NF-B IRFs AP-1 IFN Jak1 P PStats P Tyk2 P ISGs IFN can inhibit HCV replication - in subgenomic replicon models 1b (1b:Frese, M. et al, JGV, 2001; 1a: Gu et al, JV, 2003) -in genomic replicon models (1b:Blight,K. et al, Science,2000 ) -in cellular models infected -with HCVcc JFH1 (Kim,C. JVI,2007) -in primary hepatocytes infected with HCV serum (Castet et al, J Virol, 2002) - in human hepatocyte chimeric mice infected with HCVcc 1a and 2a (Hiraga, N. et al, FEBS, 2007) or with patient serum 1b and 1a (Inoue,K. et al, Hepatol. 2007) ……and in HCV chronically infected patients but efficacity of treatment not 100% Which IFN-induced gene (s) is (are) involved in the inhibition of HCV? Some ISGs and role in antiviral defense Name OAS PKR ISG56/ISG54 ISG15 ISG16 Mx Viperin NOXA Phospholipid scramblase RIG-I, MDA5 IRF7 Function RNA degradation Inhibition initiation translation Inhibition initiation translation ISGylation of proteins Inhibition of apoptosis Dynamin GTPases, bind viral nucleocapsids SAM/radical catalysis BH3-only, apoptosis Redistribution of phospholipids on Mbs; Role in Trail-induced apoptosis Boosts IFN induction Boosts IFN induction The discovery of the OAS, 2-5A and PKR in the 70s IFN-treated cells Virus- infected IFN-treated cells dsRNA Cell free Translation system Inhibition translation Hunt, T. and Ehrenfled, PNAS 1971 Cell free Translation system Inhibition translation Kerr,I. et al; Nature 1974 Inhibition of translation was dependent on IFN treatment of cells and of presence of dsRNA, produced during the viral infection Identification of two IFN-induced dsRNA-activated enzymes: -Oligoadenylate synthetases - PKR Roberts, R. et al; Nature 1976 OAS/RNase L and control of protein synthesis through the degradation of RNA The human OAS family 346 346 OAS1 OAS2 OAS3 364aa p40 I p69 342 II P56-59 OASL 400aa p46 p30 513aa 219 254aa (OAS domain but no catalytic function) Ubl domain homologous to ISG15 683 687aa 727aa p71 p100 Ubl Ubl 46% (64%) I 340 400 60% (74%) II 739 III 1087 aa 49% (66%) 44% (66%) Rebouillat &Hovanessian, 1999; Hovanessian & Justessen, 2007 OAS activates upon binding to dsRNA to generate 2’-5-linked oligoadenylates dsRNA OAS1 OAS2 Homotetramer cytosol,nuclei Donor substrate nATP NTP NTP N: A,U,C,G,I dA,dU,dG,dC 3’dA + + + 25bp the most efficient, not sequence specific OAS3 Monomer Dimer membranes Acceptor substrate ATP pppA2p5’A RpA R: tRNA A5’ppp5’A A5’pppp5’A NAD+ polyA ribosomes Product pppA(2’p5’A)n + nPPi (from dimers to 30-mers) pppA2’p5’A2’p5’N +PPii RpA2’p5’N + PPi 2-5A PPP 5’ 4’ 5’ P 5’ 1’A P A 1’ 3’ 2’ 4’ 3’ 2’ 3’ 2’ A 1’ 2-5A activates a latent Ribonuclease or RNAse L 2-5A 2 PUG STYKc dimerization RNAse L degrades RNA Degradation of cellular mRNA, rRNA, and viral RNAs UU 3’P UG 3’P UA 3’P AU 3’P 2-5A rapidly degraded by 2’-phosphodiesterase and 5’ phosphatase RNAse L KO Mice: increased sensitivity to RNA viruses (PicoRNAviridae, Reoviridae, Togaviridae, Paramyxoviridae, Orthomyxoviridae, Flaviviridae, Retroviridae) Silverman, R. JV, 2007 OAS, Rnase L and genetic predisposition to viral infections or tumors - OAS1b and susceptibility to flavivirus -The murine OAS family: 8 OAS1, 1 OAS2, 1 OAS3, 2 OAS-RL -Inbred laboratory strains susceptible to Flavivirus infection Wild mice such as Mus musculus domesticus are resistant The locus of resistance/susceptibility to Flavivirus was mapped on mouse chromosome 5 at a cluster of OAS genes Susceptibility to infection is due to a stop codon in exon 4 in the OAS1b gene leading to a truncated Inactive OAS ( Perelygin, A et al; PNAS, 2002; Mashimo et al, ; PNAS, 2002) -Rnase L and prostate cancer: 13% of patients with prostate cancer present a mutant allele of RNaseL encoding for a variant of Rnase L with substitution Arg462Gln Casey et al, 2002, Nature Genetics 40% of these patients harvour a novel gamma retrovirus (XRMV). Urisman,A. et al, PLOS Pathog.2006 The OAS/2-5A/RNase L pathway and HCV -OAS levels are elevated in HCV-infected livers and Rnase L is expressed in livers (Zhou et al, 2005) -HCV RNA are susceptible to cleavage by RNase L (in cytoplasmic extracts or with purified Rnase L) -HCV RNA ORFs are GC rich. UA and UU dinucleotides were found among the least abundant dinucleotides in HCV RNA ORFs from 162 isolates. Rnase L may exert selective pressure on HCV. (Washenberger et al, Virus res.2007). RNase L can induce IFN Transcriptome induced by 2-5A revealed several IFN-stimulated genes Malathi et al, PNAS,2005 2-5A can induce IFN in RNase L +/+ but not in RNase L -/- cells. IFN induction in response to 2-5A was greatly reduced in RIG-I and IPS1 -/- cells RNA cleavage products generated by RNase L are responsible for IFN induction Free 5’ triphosphorylated or duplex structures is thought to discriminate self (cellular) from non self (viral) RNA. Rnase L generates 3’- monophosphoryl groups from self RNA and from some viral RNA. Activation of RIG-I/Mda5 could be mediated through the duplex structure of these cleaved small RNAs. Malathi et al, Nature,2007 PKR and control of protein synthesis at the level of initiation of protein synthesis PKR Protein Kinase dsRNA dependent 1 18 80 107 159 234 272 275 551 V VI DRBD1 DRBD2 RSKKEAKNAAAKLALEIL Basic domain STKQEAKQLAAKLAYLQIL Catalytic subdomains I-XI Recognition motif for eIF2 dsRNA binding domains eIF2 eIF2-P Cytosolic serine/threonine protein kinase: -activated by dsRNAs -activated by PACT at the endogenous level -inhibited by TRBP at the endogenous level Only two known physiological substrates: - subunit of protein synthesis initiation factor eIF2 -itself PKR activation by dsRNA PKR autophosphorylation occurs in cis or through the action of a PKR dimer or a PKR monomer Back to back arrangement of the kinase domain prevents trans-autophosphorylation of each monomer within the dimer Dar et al, Cell, 2005 CONTROL OF INITIATION OF PROTEIN SYNTHESIS BY PKR e-IF2B Met-tRNA GTP eIF2-GTP GDP 60S rRNA mRNA e-IF2 -GTP-Met tRNA 40S rRNA Pre-initiation complex eIF2 -GDP CONTROL OF INITIATION OF PROTEIN SYNTHESIS BY PKR e-IF2B Met-tRNA stop eIF2 -P -GDP eIF2 -P -GDP 60S rRNA mRNA e-IF2 -GTP-Met tRNA PKR 40S rRNA preinitiation complex Shut-off of protein synthesis DOUBLE-STRANDED RNA BINDING DOMAIN (DRBD) PKR(55-79) PKR(145-168) G E G R S K K E A K N A A A K L A V E I L N K E G T G S T K Q E A K Q L A A K L A Y L Q I L S E TRBP (97-120) TRBP (227-250) G Q G P S K K A A K H K A A E V A L K H L K G G G S G T S K K L A K R N A A A K M L L R V H T V Staufen(634-658) G E G N G K K V S K K R A A E K M L V E L Q K L Staufen(770-794) G T G N S K K L A K R N A A Q A L F E L L E A V RNAseIII(203-226)G T G S S R R K A E Q A A A E Q A L K K L E L E PACT(79-102) PACT(172-195) VacciniaE3L G E G T S K K L A K H R A A E A A I N I L K A N G K G A S K K Q A K R N A A E K F L A K F S N I A D G K S K R D A K N N A A K L A V D K L L G Y TRBP and E3L are inhibitors of PKR PACT is an activator of PKR The eIF2-KINASE family ACTIVATION UNDER DIFFERENT CONDITIONS OF STRESS HRI GCN2 PKR PERK Heme levels oxydative stress Heat shock Amino acid Deprivation Ds RNA ER stress Misfolded proteins Heme, Hsp90, Hsc70 Uncharged tRNA dsRNA Ire1 motifs HisRS-related eIF2-P eIF2-P eIF2-P RE eIF2-P DRBD and binding to nucleic acids -topology ---- -interaction with dsRNA through contact with OH-groups and with phosphates --no interaction with dsDNA or RNA/DNA -no recognition of specific sequences -The DRBD-containing proteins do not bind dsRNA similarly: differents contact sites, different dsRNA lengths Inactive Active -Non specific binding of PKR to 20 bp dsRNA. No activation -Specific binding of PKR to 30 bp. Binding of two PKR. Autophosphorylation and active. The role of dsRNA is to bring 2 or more PKR monomers in close proximity to enhance dimerization via the kinase domain.(Lemaire,P. et al, JMB,2008) ISG54/ISG56 and control of protein synthesis at the level of eIF3 eIF3 and initiation of protein synthesis Translation CAP dependent: eIF3 binds 40S and recruits m7GpppRNA via interaction with eIF4F (eIF4E, eIF4A, eIF4G) Translation CAP independent: eIF3 (10-12 proteins) binds directly to IRES Similarity of HCV IRES and eIF4F to anchor the mRNA Strand near the exit site of the 40S rRNA HCV does not need eIF4F for viral protein synthesis S13 40S-eIF3-IRES model 40S-eIF3-eIF4F-mRNA model Siridechadilok,B.et al, Science, 2005 ISG56 and ISG54 and control of translation -Belongs to the early ISGs: induced by IFN and rapidly induced by viral stresses Related (42% sequence conservation); contain TPR motifs CAP-dependent initiation of translation ISG56 (2 IRF3 and one NF-B binding sites) ISG54 (2 IRF3 binding sites only) Both bind initiation factor eIF3: ISG54 binds subunit c and e ISG56 binds subunit e Both block formation of stable eIF3-ternary complex In addition, ISG54 interferes with formation of preInitiation complex ISG56 could inhibit HCV translation through interaction with eIF3 S13 Wang,C. et al, JVI, 2003 Terenzi,F. et al, JBC, 2006 ISG15 and post-translational modification of proteins ISG15 30% homology to Ubiquitin 30% homology to Ubiquitin LRLRGG Conjugation to Cysteine Residues of conjugating enzymes UBCH6( UBE2E1) and UBCH8 (UbE2L6) (HERC5, TRIM25) (USp18(UBP43),USP2,USP5,USP13,USP14) Mechanism of action of ISG15 Activation effect: Modification of Components of the host immune Response ISG15 and all its conjugated and deconjugated enzymes are IFN-induced genes Targets of ISG15: ≈160 genes (examples :JAK1, STAT1,PRRs (RIG-I), IRF3, MxA, PKR, RNase L) ISG15 can increase IFN induction (prevents degradation of IRF3) ISG15 KO mice:increased susceptibility to infection (flu, Sindbis, HSV-1) ISGylation of HIV-1 Gag and Tsg101 Inhibits their ubiquitination (required for virion release) USP18 KO mice more resistant to viral Infections (LCMV, VSV) Sadler, A and Williams, B. Nature reviews, 2008 HCV interferes with multiple cellular pathways Binds STAT3,binds Smad3 provokes cellular proliferation Increases mitochondrial Ca2+ uptake Inhibits IFN induction Associates with PKR,Grb2,p53 Inhibits ISG induction through induction of IL-8 Binds and inhibits Rb Stimulates S phase entry Role intransformation? Associates with PKR and PERK HCV NS5A Domain I 1-213 Domain II 250-342 Domain III 256-447 447 Zinc binding domains Amphipathic helix Anchors NS5A to the ER Cluster of hyperphosphorylation Polyproline cluster NLS ISDR heterogeneity of sequence associated with sensitivity or resistance to IFN treatment (genotype 1b) Enomoto N, N.Engl.J.Med 1996 The maintenance or evolution of the Domain II or Domain D2 ISDR sequence is linked to its role -is intrinsically unfolded In the viral life cycle and the progression -interacts with NS5B of liver disease -contains the ISDR -contains a PKR-binding domain -contains a Bcl-2 homology domain -interacts with GRB2, with the death domain of Myd88 -activates PI3kinase by binding to an SH3 domain of one regulatory subunit -Provokes induction of IL-8 which then inhibits induction of ISGs NS5A involved in replication and pathogenesis VIRAL INHIBITORS OF PKR Competition for dsRNA E3L (VV) 3 and 4 (Réovirus) NS1 (Influenza) NSP3 (rotavirus) TRS1,IRS1 (HCMV) Degradation Poliovirus-activated protease Compartimentalization EMCV Sequestration by dsRNA EBERs (EBV), VA1 (Adénovirus) NH2 NH2 Direct interaction p58 IPK (cellular protein activated by influenza) Tat (HIV-1) soluble E2 (HCV) cIRF2(KSHV) SM (EBV) NS5A (HCV) Us11protein (HSV1) Competition With substrate K3L (Vaccinia) Substrat Inhibition of eIf2 phospho LANA2 (KSHV) Substrat-P Activation of phosphatase 1 1 34.5 Kda (HSV1) HCV E2 Hypervariable region Signal leader PePHD (276-287) TM 1 2 363 HCV 1a/b R S E L S P L L L T T T 18 80 107 79- 90 PKR 48-54 eIF2 159 PKR 1 234 272 KKAVSPLLLTTT SELSRRS 275 551 V VI DRBD1 DRBD2 51 79-83 eIF2 KGYID Taylor et al,1999 Catalytic subdomains I-XI 314 INHIBITION OF PKR BY HCV E2 A cytosolic non-glycosylated domain of E2 binds to and inhibits PKR (and PERK) Restoration of translation Fractions 1 2 3 4 5 Glycoprotein 68 p38 E2 glycosylated top Bottom ***** E1 *********** E2 ® ER lumen PERK Pavio et al. J. Virol. 2003 ® Cytosol E2 E2 non glycosylated PKR AAAAA Pavio et al. J. Virol. 2002 Is there a correlation with mutations in NS5A ISDR and in E2 PePHD and with IFN treatment outcome? 4 mutations at least in NS5A ISDR Genotype 1b Genotype 1a/b Sustained virologic response to IFN- Japan, Western C. Genotype 2a Genotype 3 mutations outside the NS5A ISDR Genotype 1b Genotype 1a Mutations in E2 PePHD Genotype 1b Genotype 3a Genotype 2a/b yes Yes, but only a small number of mutations yes no Sustained virologic response to IFN- In the carboxy terminal part of NS5A, in the Variable region 3 and flanking regions Sustained virologic response to IFN- no no Conflicting reports Hofmann, W. et al, J Clinical Virol, 2005 inhibition of PKR activity by a pseudosubstrate: the Vaccinia K3L protein Ser51 314 79-83 human eIF-2 Several binding motifs for PKR KGYID 1 vaccinia K3L 33% identity and 68% similarity with eIF2- K 88 KGYID essentiel binding motif to PKR 1 K 71 K3L322 mutant KGAID Stronger PKR inhibitor than K3L He, cell Death and diff.2006; Garcia et al, Biochimie, 2007 OTHER WAYS TO ESCAPE CONTROL OF TRANSLATION BY PKR -Alphavirus (Sindbis virus, Semliki Forest Virus): positive ssRNA Translation of subgenomic 26s RNA efficient in presence of PKR and eIF2-P hairpin loop structure (DLP) : stalls scanning and allows efficient presentation of Mtet tRNA to AUGi through an other factor: eIF2A, in the absence of eIF2 Ventoso, I. Genes & Development, 2006) PKR KO ( e-IF2 functional ) Met tRNA GTP PKR +/+ ( e-IF2 non functional ) Met tRNA Met tRNA e-IF2 e-IF2 AUGi eIF2A DLP DLP AUGi scanning scanning efficient DLP DLP Met tRNA GTP e-IF2 Met tRNA Met tRNA Met tRNA e-IF2 AUGi scanning efficient Met tRNA e-IF2 AUG inefficient AUGi scanning Very inefficient AUG Very inefficient How does Interferon work on HCV? Several ISG are detected in chronic Hepatitis . This reveals an ongoing response to endogenous IFN and/or viral RNA (Helbig, K. et al, Hepatol.2005) Which genes are linked to HCV clearance in vivo? Decrease of viral levels depends on the interplay of several ISGs RIG-I, MDA5 : provokes IFN induction OAS and RNase L : -provokes degradation of viral RNA and also sustain IFN induction ( through RIG-I activation) PKR and ISG56 Viperin : inhibit initiation of translation on eIF2 and eIF3 : interference with protein assembly at the ER? STAT1 : sustains IFN induction ISG15, USP18 : ISGylation, modification of trafficking NOXA : apoptose genes hypothesized to be important in the anti-HCV response RIG-1 eIF2 ISG16: inhibitor of apoptosis Ubiquitin ligase Novel mathematical method used to explore association between decrease in viral titers and change in gene expression in HCV patients following PegIFN/Ribavirin treatment Brodsky,L. et al PLoS one, 2006 ISG56 IRF7 Mx1 and 2 OAS 69/71 kDa OASL VIPERIN USP18 Factors involved in the persistence of HCV in the host -Genetic variability of the virus; lack of fidelity of NS5B (1,5-2x10-3 mutation/ nucleotide/year), no proofreading (no 3’-5’ exonuclease activity).Generation of escape mutants -mutations in HVR1 and 2 of E2 (weakens humoral defense) -mutations in NS3/7A protease (resistance to the new anti-protease VX-950, SCH 503034) -Host factors: -cellular immune response weak or defective ; -Overreaction of cytotoxic T cells and NK cells leading to destruction of infected hepatocytes And production of inflammatory cytokines provoking liver damages -Induction of endogenous IFN compromised by inhibition of its induction pathway through the NS3/4A protease Why IFN treatment is not 100% efficient on HCV infection? Interference of the virus with cellular signalling pathways -NS3/4A prevents sustained induction of endogenous IFN -through RIG-I/MAVS by cleaving MAVS -through TLR3/TRIF by cleaving TRIF -Induction of SOCS proteins interfere with STAT function -NS5A and cytosolic E2 interfere with PKR -core interacts with STAT1, provokes its degradation by proteasome -NS5A provokes induction of IL-8 through NF-B; IL-8 inhibits IFN action