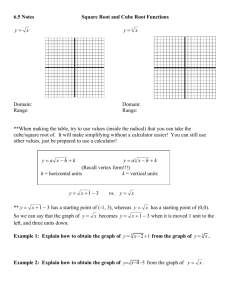
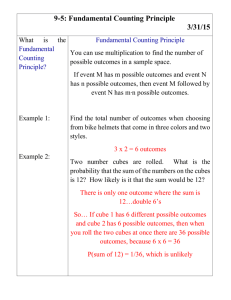
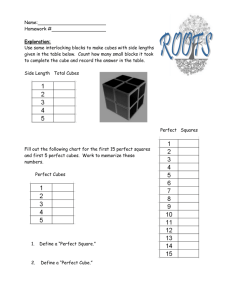
Mark Blackburn
advertisement

Marching Cubes: A High Resolution 3D Surface Construction Algorithm William E. Lorenson Harvey E. Cline General Electric Company Corporate Research and Development, SIGGRAPH 1987 Presented by Mark Blackburn, Fall 2005 ●●●●●●●●●●●●●●●●●●●●●●●●● Organization 1. 2. 3. 4. 5. 6. 7. Motivation Related Work Information Flow Algorithm Short Demo Results Conclusion & Future Work ●●●●●●●●●●●●●●●●●●●●●●●●● Motivation • 3D visualization is critical to medical field • Existing algorithms inadequate: – lack detail – may introduce artifacts • Create polygonal representation of constant density surfaces from 3D array of data ●●●●●●●●●●●●●●●●●●●●●●●●● Related Work • Existing methods of 3D surface generation – Trace contours within each slice then connect with triangles ( cf. topography map) – Create surfaces from “cuberilles” (voxels) – Ray casting to find the 3D surface • Use hue-lightness to shade surface • Use gradient to shade – Display density volumes ●●●●●●●●●●●●●●●●●●●●●●●●● Related Work Shortcomings of Existing Techniques • Throw away useful information in the original data – Cuberilles: uses thresholding to represent surface – Ray casting: uses depth shading alone or approximates shading using unnormalized gradient • Some lack hidden surface removal – Volume models display all values and rely on motion to produce a 3D sensation ●●●●●●●●●●●●●●●●●●●●●●●●● Related Work • Benefits of Marching Cubes approach: – Uses all information from source data – Derives inter-slice connectivity, surface location, and surface gradient – Result can be displayed on conventional graphics display systems using standard rendering algorithms ●●●●●●●●●●●●●●●●●●●●●●●●● Information Flow Information Flow for 3D Medical Algorithms – – – – Data Acquisition : collect a series of 2D slices of information Image Processing: find structures with 3D data or filter / bin data Surface Construction: Create surface model of voxels or polygons Viewing & Display: Display the surface using ray casting, depth or color shading. ●●●●●●●●●●●●●●●●●●●●●●●●● Algorithm Overview • The Marching Cubes Algorithm Consists of 3 basic steps: 1. Locate the surface corresponding to a userspecified value. 2. Create triangles. 3. Calculate normals to the surface at each vertex. ●●●●●●●●●●●●●●●●●●●●●●●●● Step 1:Surface Intersection • To locate the surface, it uses a logical cube created from eight pixels (Four each from 2 adjacent layers): ●●●●●●●●●●●●●●●●●●●●●●●●● Step 1: Surface Intersection • Binary vertex assignment: (p (i, j, k) >= TU ? 1: 0) – Set cube vertex to value of 1 if the data value at that vertex exceeds (or equals) the value of the surface we are constructing – Otherwise, set cube vertex to 0 • If a vertex = 1 then it is “inside” the surface • If a vertex = 0 then it is “outside” • Any cube with vertices of both types is “intersected” by the surface. ●●●●●●●●●●●●●●●●●●●●●●●●● Step 2 : Triangulation • For each cube, we have 8 vertices with 2 possible states each (inside or outside). • This gives us 28 possible patterns = 256 cases. • Enumerate cases to create a LUT • Use symmetries to reduce problem from 256 to 15 cases. ●●●●●●●●●●●●●●●●●●●●●●●●● Step 2 : Triangulation • Use vertex bit mask to create an index for each case based on the state of the vertexes. • Using the index to tell which edge the surface intersects, we can then can linearly interpolate the surface intersection along the edge. • In our previous example of green pixels, v0 and v3 were “outside” the surface and thus our index would = 11110110 = 246 ●●●●●●●●●●●●●●●●●●●●●●●●● Step 3 : Surface normals • • • To calculate surface normal, we need to determine gradient vector, g (derivative of the density function). To estimate the gradient vector at the surface of interest, we first estimate the gradient vectors at the vertices and interpolate the gradient at the intersection. The gradient at cube vertex (i , j, k), is estimated using central differences along the three coordinate axes by: D (i, j, k) is the density at pixel (i, j) in slice k. Δx, Δy, Δz are lengths of the cube edges ●●●●●●●●●●●●●●●●●●●●●●●●● Step 3 : Surface normals • Dividing the gradient by its length produces the unit normal at the vertex required for rendering. • Then the algorithm linearly interpolates this normal to the point of intersection. ●●●●●●●●●●●●●●●●●●●●●●●●● Algorithm Summary 1. 2. 3. 4. 5. Scan 2 slices and create cube Calculate index for cube based on vertices Use index to lookup list of edges intersected Use densities to interpolate edge intersections Calculate unit normal at each edge vertex using central differences. Interpolate normal to each triangle vertex 6. Output the triangle vertices and vertex normals 7. March to next position and repeat. ●●●●●●●●●●●●●●●●●●●●●●●●● Demo • Short Demo / tutorial : http://www.essi.fr/~lingrand/MarchingCubes/applet.html ●●●●●●●●●●●●●●●●●●●●●●●●● Enhancements • Efficiency – Take advantage of pixel-to-pixel, line-to-line, and slice-to-slice coherence by keeping previous calculations. • Functional – Added solid modeling capability • Boolean operations permit cutting and capping of solid models as well as multiple surface extraction. ●●●●●●●●●●●●●●●●●●●●●●●●● Results Bone Surface Soft Tissue Surface ●●●●●●●●●●●●●●●●●●●●●●●●● Results – The Visible Human Inside skeleton view Torso / bowels Images courtesy : Marching Through the Visible Man: http://www.essi.fr/~lingrand/MarchingCubes/accueil.html) ●●●●●●●●●●●●●●●●●●●●●●●●● Results –The Visible Man ●●●●●●●●●●●●●●●●●●●●●●●●● Results Human brain surface reconstructed by using marching cubes (128,984 vertices and 258,004 triangles Magnified display of brain surface Images courtesy : IF of Polytech' Nice-Sophia: http://www.essi.fr/~lingrand/MarchingCubes/accueil.html) ●●●●●●●●●●●●●●●●●●●●●●●●● Conclusion • Marching Cubes is an algorithm for 3D surface reconstruction well suited to series of 2D medical images (slices). • Algorithm summary ( 3 steps) 1. Locate the surface corresponding to a user-specified value using cube vertex tests 2. Create triangles based on vertex states of the cube 3. Calculate normals to the surface at each vertex. ●●●●●●●●●●●●●●●●●●●●●●●●● Future Work • Dividing Cubes: – An algorithm that generates points rather than triangles. As the resolution of the data increases, the number of triangles approaches the number of pixels. • Dual Marching Cubes – creates isosurfaces very similar to Marching cubes surfaces, but they are comprised of quad patches that eliminate some of the problems of poorly shaped triangles that can occur in this algorithm. ●●●●●●●●●●●●●●●●●●●●●●●●● Evaluation • Advantages: • Uses all information from source data • Derives inter-slice connectivity, surface location, and surface gradient • Result can be displayed on conventional graphics display systems using standard rendering algorithms • Allows Solid modeling capability ( cutting and capping) • Disadvantages: • Requires user input • Mainly limited to medical images with clear contiguous intensity boundaries (constant density) • Is performing a modified form of thresholding. ●●●●●●●●●●●●●●●●●●●●●●●●● References • W. Lorensen and H. Cline. “Marching cubes: A High Resolution 3D Surface Construction Algorithm”, Proceedings of SIGGRAPH 1987, pages 163-169, 1987. • The Visible Human: http://www.nlm.nih.gov/research/visible/visible_human.html • Marching Cubes Demo/Tutorial: http://www.essi.fr/~lingrand/MarchingCubes/applet.html • Nielson, Gregory M. “Dual Marching Cubes”, IEEE Visualization archive Proceedings of the conference on Visualization '04, Pages: 489 - 496