60ºC 35 cycles - Naresuan University
advertisement

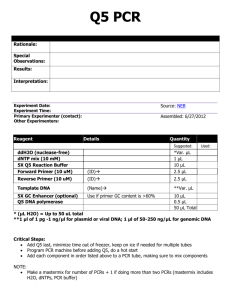
DEVELOPMENT OF TETRA-PRIMER ARMS-PCR FOR HEMOGLOBIN E DETECTION Jatuphol Pholtaisong Biological Science Program Department of Biology, Faculty of Science, Naresuan University Advisor: Dr.Lt.Saisiri Mirasena Co-Advisors: Asst.Prof.Dr.Maliwan Nakkuntod and Dr.Phrutthinun Surit Hemoglobin E Abnormal Hemoglobin -globin gene Codon 26 (GAG-AAG) Glu-Lys Alternative splicing => reduced mRNA Mildly unstable (sensitive to oxidants) Beta thalassemia/Hemoglobin E => Mild to Severe Lower Northern Part of Thailand (Rawangkran et al.,2013) - Homozygous HbE 5.05% - Heterozygous HbE 42.09% 2 Hemoglobin E Detections Screening Tests - Dichlorophenol indophenol (DCIP) - High-Performance Liquid Chromatography (HPLC) Confirmation Tests - Reverse Dot-Blot Hybridization - Amplification Refractory Mutation System–PCR (ARMS-PCR) 3 Amplification Refractory Mutation System–PCR (ARMS-PCR) Forward Primer 5’ 3’ Reverse Primer (Normal) 3’ C 5’ Reverse Primer (Mutant) 3’ T 5’ Reaction 1 Reaction 2 Normal Allele Detection Mutant Allele Detection Forward Primer 5’ 3’ Forward Primer 5’ 3’ Reverse Primer (Normal) 3’ C 5’ Reverse Primer (Mutant) 3’ T 5’ 4 Amplification Refractory Mutation System–PCR (ARMS-PCR) Interpretation 1 (N) 2 (M) Normal 1 (N) 2 (M) Heterozygous HbE 1 (N) 2 (M) Homozygous HbE 5 Amplification Refractory Mutation System–PCR (ARMS-PCR) Interpretation 1 (N) 2 (M) 1 (N) 2 (M) 1 (N) 2 (M) Internal > Control Normal Heterozygous HbE Homozygous HbE 6 Tetra-primer ARMS-PCR Principle Ye et al. (2001) 7 Primer Mismatch at 3’-end 5’ G 3’C 5’ G 3’T Mismatch 5’ 5’ 3’ 3’ 8 Weak Primer Mismatch at 3’-end 5’ G 3’C 5’ G 3’T Weak Mismatch 5’ 5’ 3’ 3’ 9 Type of Primer Mismatch Strong Mismatch Medium Mismatch Weak Mismatch G/A C/T A/A C/C G/G T/T C/A G/T > Add secondary mismatch at position –2 from the 3’-terminus to increase specificity Ye et al. (2001) 10 Study Steps Primer Design PCR Optimization Compare with ARMS-PCR 11 Tetra-primer ARMS-PCR Principle Ye et al. (2001) 12 Materials and Methods: Primer Design PRIMER1: primer design for tetra-primer ARMS-PCR (Ye et al., 2001) OligoCalc (Kibbe, 2007) OligoAnalyzer 3.1 (https://sg.idtdna.com/calc/analyzer) GenBank (http://www.ncbi.nlm.nih.gov/genbank/) Primers Sequences TM (ºC) Position Product size HbE-OF CCC TTC CTA TGA CAT GAA CTT AAC CAT A 65.6 5226506-5226533 (NC_000011.10) 691 HbE-OR GGC TGT CAT CAC TTA GAC CTC AC 64.6 5227174-5227196 (NC_000011.10) HbE-IF(Normal) 64.6 5226921-5226943 (NC_000011.10) 276 ACCAACCTGCCCAGGGCATC HbE-IR(Mutant) GTGAACGTGGATGAAGTTGGTGTTA 64.1 5226943-5226967(NC_000011.10) 462 13 Study Steps Primer Design PCR Optimization Compare with ARMS-PCR 14 PCR Optimization: HbE Samples 15 PCR Optimization: HbE Samples 16 PCR Optimization: HbE Samples 17 PCR Optimization Primer Concentration 0.2 µM Each Primer 0.2 µM OF/OR Primer, 0.4 µM IF/IR Primer Annealing Temperature Normal Annealing Temperature (60ºC 35 cycles) Touchdown PCR - 70ºc (-1ºc/cycle) 10 cycles - 60ºc 25 cycles 18 Results: PCR Optimization 0.2 µM Each Primer Normal Annealing Temperature (60ºC 35 cycles) M: Marker 1: Normal 2: Heterozygous Hb E 3: Homozygous HbE 4: Negative 19 Results: PCR Optimization 0.2 µM Each Primer 0.2 µM OF/OR Primer, 0.4 µM IF/IR Primer Normal Annealing Temperature (60ºC 35 cycles) M: Marker 1: Normal 2: Heterozygous Hb E 3: Homozygous HbE 4: Negative 20 Results: PCR Optimization 0.2 µM Each Primer 0.2 µM OF/OR Primer, 0.4 µM IF/IR Primer Normal Annealing Temperature (60ºC 35 cycles) Touchdown PCR - 70ºc (-1ºc/cycle) 10 cycles - 60ºc 25 cycles M: Marker 1: Normal 2: Heterozygous Hb E 3: Homozygous HbE 4: Negative 21 Results: PCR Optimization 0.2 µM Each Primer 0.2 µM OF/OR Primer, 0.4 µM IF/IR Primer Normal Annealing Temperature (60ºC 35 cycles) Touchdown PCR - 70ºc (-1ºc/cycle) 10 cycles - 60ºc 25 cycles M: Marker 1: Normal 2: Heterozygous Hb E 3: Homozygous HbE 4: Negative 22 Results: PCR Optimization 0.2 µM Each Primer 0.2 µM OF/OR Primer, 0.4 µM IF/IR Primer Normal Annealing Temperature (60ºC 35 cycles) Touchdown PCR - 70ºc (-1ºc/cycle) 10 cycles - 60ºc 25 cycles M: Marker 1: Normal 2: Heterozygous Hb E 3: Homozygous HbE 4: Negative 23 Results: PCR Ingredients PCR Ingredients (total volume 20 µL) 1X PCR Buffer (NanoHelix, Korea) 0.2 mM Each DNTPs (NanoHelix, Korea) 1 Unit HelixAmpTM Ab+ Taq DNA polymerase (NanoHelix, Korea) 0.2 µM HbE-OF Primer, 0.2 µM HbE-OR Primer 0.3 µM HbE-IF Primer, 0.3 µM HbE-IR Primer DNA Template 3 µL 24 Results: PCR Cycles Pre-Denaturation Denaturation Annealing Extension Denaturation Annealing Extension Final Extension 95ºc 2m 97ºc 30s 70ºc (-1ºc/cycle) 30s 10 cycles 72ºc 1m 97ºc 30s 60ºc 30s 25 cycles 72ºc 1m 72ºc 5m 25 Methods: Step by Step Primer Design PCR Optimization Compare with ARMS-PCR 26 Results: Tetra Primer ARMS-PCR and ARMSPCR Comparison Known Samples: Normal 20 Samples Heterozygous Hb E 16 Samples Homozygous Hb E 20 Samples E E EE E N N E EE EE E E - 27 Discussions and Conclusions Tetra-Primer ARMS-PCR for Hemoglobin E Detection was successfully developed. This technique required touchdown PCR and higher inner primer concentrations. When compared with standard technique in 48 known samples, the results were concordant. This technique is efficient, less time-consuming and safe cost for hemoglobin E gene detection. 28 References Rawangkran, A., Janwithee, N., Wong, P., & Jermnim, N. (2013). Prevalence of Thalassemia Trait from Screening Program in Pregnant Women in the Lower Northern Region of Thailand. Thai Journal of Genetics, S(1), 156-159. Ye, S., Dhillon, S., Ke, X., Collins, A. R., & Day, I. N. (2001). An efficient procedure for genotyping single nucleotide polymorphisms . Nucleic Acids Research, 29(17), E88-88. doi: 10.1093/nar/29.17.e88. 29 Acknowledgments 30 T G 31