Genes
advertisement

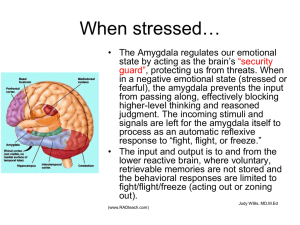
Functional Neuroimaging of Genetically Driven Variation in Brain Function: Towards a Biological Understanding of Individual Differences in Behavior Ahmad R. Hariri, Ph.D. Developmental Imaging Genomics Program Department of Psychiatry University of Pittsburgh School of Medicine Why study genes? • Various aspects of cognition, temperament, and personality are highly heritable (40-70%) • Account for the lionshare of susceptibility to major psychiatric disorders • Transcend phenomenological diagnosis, and represent mechanisms of disease • Offer the potential to identify at-risk individuals and biological pathways for the development of new treatments Deshaies 02 — DNA Man #1 How do we get from here to there? Behavior: complex functional interactions and emergent phenomena Genes: multiple susceptibility alleles each of small effect The path from here to there… IMAGING GENOMICS Behavior: complex functional interactions and emergent phenomena Genes: multiple susceptibility alleles each of small effect Cells: Subtle molecular alterations Systems: response bias to environmental cues Imaging Genomics: Basic Principles Imaging Genomics: Basic Principles 1) Selection of candidate genes – Well defined functional polymorphisms, associated with specific physiological effects at the cellular level in distinct brain circuits – Genes with identified SNPs or other allele variants with likely functional implications involving circumscribed neuroanatomical systems Imaging Genomics: Basic Principles 2) Control for non-genetic factors – Systematic differences between genotype groups could either obscure a true gene effect or masquerade for one • Age, gender, IQ, population stratification • Environmental factors such as illness, injury, or substance abuse • Task performance – Linked pari passu with BOLD response – Match or consider variability Imaging Genomics: Basic Principles 3) Task selection – Imaging tasks must maximize sensitivity and inferential value, as the interpretation of potential gene effects depends on the validity of the information processing paradigm • Engage circumscribed brain circuits • Produce robust signals in all subjects • Show variance across subjects Imaging Genomics: Applications Central serotonergic system Slide courtesy of K.P. Lesch Typical 5-HT neuron and target synapse Figure courtesy of K.P. Lesch 5-HT Transporter Promoter Variant (5-HTTLPR) Figure courtesy of K.P. Lesch The 5-HTTLPR Harm avoidance, Neuroticism, Depression, Anxiety Genes: Short and long allele variants Cells: Alterations in synaptic 5-HT 5-HTTLPR and temperament IMAGING GENOMICS Harm avoidance, Neuroticism, Depression, Anxiety Genes: Short and long allele variants Cells: Alterations in synaptic 5-HT Systems: amygdala bias to environmental cues The Amygdala fMRI amygdala reactivity paradigm (A.K.A. Hariri’s Hammer) 5-HTTLPR S allele driven amygdala hyper-reactivity to environmental cues P < 0.05, corrected Hariri et al., Science 2002 S allele driven amygdala hyper-reactivity Berlin replication in healthy adults R=0.6, p<0.005 LL LS SS Heinz et al., Nature Neuroscience 2005 Italian replication in healthy adults S carriers > L/L % Signal Change in Amygdala 0,30 0,25 0,20 0,15 0,10 0,05 0,00 ss ls ll SERT genotype P < 0.05, corrected Bertolino et al., Biological Psychiatry 2005 Pittsburgh replication in healthy adults 5-HTTLPR S carrier > LL (P < 0.05, uncorrected) Sample Demographics: LL: 8♀/4♂; Mean age = 46.1 S carrier: 9♀/7♂; Mean age 47.5 Swedish replication in social phobics Furmark et al., Neuroscience Letters 2004 Mean +/- 1 SEM Right Amygdala BOLD NIMH replication in healthy adults N = 92 .3 .2 .1 0.0 -.1 N= 27 65 L/L S Carrier 5-HTTLPR Hariri et al., Archives (2005) Elaboration: S allele load and sex effects .3 .2 .1 SEX 0.0 Female -.1 N= Male 14 13 L/L 28 23 L/S 5 9 S/S 5-HTTLPR Hariri et al., Archives (2005) 5-HTTLPR and temperament IMAGING GENOMICS ????????? Genes: Short allele variant Cells: Increased synaptic 5-HT Systems: amygdala bias to environmental cues Amygdala reactivity and harm avoidance 30 20 20 Total Harm Avoidance 30 10 10 0 0 -10 -.6 -.4 -.2 -.0 .2 Left Amygdala BOLD .4 .6 .8 -10 -.8 -.6 -.4 -.2 0.0 .2 .4 .6 Right Amygdala BOLD * No correlation between amygdala reactivity and HA .8 Prefrontal-Amygdala Dynamics Wood & Grafman 2003 Reduced functional coupling of the amygdala and prefrontal cortex in S allele carriers Overall Coupling 5-HTTLPR Effects left right Pezawas et al. Nature Neuroscience 2005 Amygdala-Prefrontal connectivity predicts HA Functional circuitry is key for understanding complex emergent phenomena 5-HTTLPR biases corticolimbic information processing related to temperament Hamann Nature Neuroscience 2005 Subgenual PFC 5-HT1A and 5-HT2A binding predict amygdala reactivity sgPFC 1A/2A ratio predicts amygdala reactivity DRN 5-HT1A predicts amygdala reactivity Typical 5-HT neuron and target synapse Figure courtesy of K.P. Lesch hTPH2 G(-844)T polymorphism • Relatively high minor allele frequency (T allele = 38%) • Located within 1 Kb (844 bp upstream) of the transcription initiation site of hTPH2 and is likely a constituent of the proximal promoter of the gene • Regulatory variants often produce functional changes in gene expression • Transcriptional regulatory databases indicate transcription factor recognition sequence homology surrounding the -844 promoter variant (http://www.genomatix.de) • In silico evidence that the G to T allele substitution potentially modifies the binding of several transcription factors including octamer-binding factor 6, special AT-rich sequence-binding protein 1 as well as homeodomain proteins MSX-1 and MSX-2 hTPH2 G(-844)T biases amygdala reactivity Right amygdala activity (in arbitrary units) 2.00 1.00 0.00 -1.00 T carriers > G/G G/G T carrier hTPH2 genotype Brown et al., Molecular Psychiatry (in press) hTPH2 G(-844)T biases amygdala reactivity Emotional Behaviors? Genes: hTPH2 expression? Cells: 5-HT synthesis? Systems: amygdala bias to environmental cues Acknowledgments " " University of Pittsburgh Steve Manuck Bob Ferrell Carolyn Meltzer Sarah Brown Patrick Fisher Scott Kurdilla NIMH - GeCaP Danny Weinberger Emily Drabant Karen Munoz Anand Mattay Lukas Pezawas Andreas Meyer-Lindenberg Support: NIMH P01MH041712-18, R24MH067346-03, R01MH061596-04; NIDA R01DA018910-01; NARSAD