What is Happening in Peanut Genomics in India
advertisement

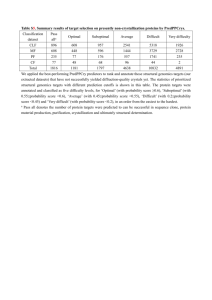
What is happening in peanut genomics in India Rajeev Varshney Senior Scientist- Applied Genomics, ICRISAT Coordinator, Indian Groundnut Genomics Consortium Leader, Comparative Genomics & Gene Discovery (Generation Challenge Programme) Groundnut production in India • 1st most important oilseed crop in India Area: 6.6 million ha Production: 5.9 million t Productivity of groundnut – 0.97 t ha-1 ~ 20% of world production comes from India alone! Constraints in production Drought Leaf spot Rust Indian Groundnut Genomics Consortium - Indian Council of Agricultural Research Three main partners: ICRISAT, Patancheru, A.P. (Intern. Crops Res. Inst. Semi-Arid Tropics) NRCG, Junagadh, Gujarat (National Research Centre for Groundnut) UAS, Dharwad, Karnataka (University of Agricultural Sciences) Team of multidisciplinary scientists Activities • Generation of microsatellite-enriched libraries and identification of ~500 microsatellite or simple sequence repeat (SSR) loci • Generation of groundnut unigene-derived SNP markers and development of cost effective CAPS assay • Construction of integrated genetic maps with SSR, SNP and DArT markers • Phenotyping of mapping populations at three locations (ICRISAT, NRCG, UASD) over 3 years • Identification of genes/QTLs associated with resistance to foliar diseases (rust and late leaf spot) and tolerance to drought (TE, SCMR and SLA) What we already have done at ICRISAT… SSR marker development In silico approach after mining sequence data - Aeschynomenoid/ Dalbergoid and Genistoid clades of the Leguminoae (15 genera) - 6 genera Adesmia, Amorpha, Dalbergia, Chaetocalyx Lupinus and Stylosanthes: 411 seqs contained SSRs . - Primer pairs for 109 unique SSRs - 79 functional in one of six genera - 51 primer pairs functional in peanut BAC-end sequence derived SSRs (Doug Cook) Genetic map construction Ah1 Ah2 0.0 GM635a 5.9 GM635b seq18G09_5B seq13A07 GM669 seq17E01 10.7 11.6 14.5 19.0 0.0 3.3 6.2 7.6 9.4 14.5 Ah3 GM630 seq10D04 RN6F05 GM633 PM179 TC6E01 GM626 0.0 seq16C06 15.5 seq13E09 30.3 42.6 Ah10 seq2B09 GM672 TC2D06 GM667 Ah5 TC3G01b 0.0 XIP219 XIP606 XIP419 XIP123 PM721 GM660 GM679 XIP177 XIP406 TC9F10_8A TC6H03_6B PM733a 18.0 24.3 26.5 28.1 29.2 29.8 31.5 33.8 35.9 37.4 39.2 40.9 TC3G01a TC3G01c 48.4 50.1 Ah9.2 Ah4 0.0 13.9 Ah11 XIP287 0.0 Ah12 TC11B04 23.6 27.1 seq19D06 37.3 GM723b 43.7 seq19H03 GM647 0.0 TC7C06 11.0 XIP245 16.4 18.9 22.6 GM623 TC1A02_7B XIP689 28.4 TC1A01 34.8 PM377 38.6 seq9H08a 46.1 XIP171A Ah15 Ah14 TC4D09 0.0 PM427 0.0 XIP105 PM375 GM629 GM736 XIP108A XIP524 7.7 10.8 13.2 6.7 10.3 22.5 24.6 seq19A05 XIP165 23.9 26.1 seq18C05 XIP108B Ah8 Ah7 0.0 RI1F06_2A 8.6 seq18E07 17.1 18.9 seq1B09 PM499 25.3 PM73 34.6 AC2C05 0.0 seq18G01 9.8 seq19D01 0.0 22.5 50.8 seq2G04 62.0 TC9F04 Ah16 0.0 seq9G05 Ah193_1A Ah6 0.0 Ah17 TC4G02 Ah9.1 GM698 seq8H01 Ah18 0.0 GM624 5.4 9.0 seq19B12 TC9H09 20.1 seq15C10 0.0 3.8 0.0 seq7G02 18.6 20.7 22.7 26.8 28.8 gi1170_9A Lec1_9A PM436 GM694 seq14H06 34.5 TC5A06 Ah19 seq11G03 TC4F12_2A 0.0 Ah9.2 Ah10 0.0 seq2B09 0.0 XIP 14.6 16.2 GM672 TC2D06 13.9 seq 23.6 Ah193_1A 27.1 seq 37.3 GM 43.7 seq 61.8 PM 75.6 PM 32.9 seq13A10 Ah20 seq19D09_1B 0.0 GM641 seq13A10 42.1 36.7 GM618 51.9 Gi4926 62.9 GM745 67.5 seq18A05 83.0 seq19G07 35.4 TC7E04 TC7H11_2A Collab. D Bertioli, Brazil G He, USA S Knapp, USA 47.3 61.8 75.6 PM183 60.8 GM692 61.1 XIP136 PM418 Cultivated x Cultivated TAG 24 x ICGV 86031 -RIL seq16G08 Mapping water use efficiency QTLs Molecular diversity 916 accessions at 21 SSR loci fastigiata: 488 hypogaea: 321 wild: 47 Reference collection (300 genotypes) Exploring association genetics approach What we plan to do at ICRISAT (under Troplical Legume Genomics project)… Generation Challenge Programme (www.generationcp.org) • Launched in August 2003 • 10-year framework (2004–2008; 2009–2013) • About US$16M annual budget – 75% Research – 15% Training and capacity building – 10% Management cost • Major donors – European Community – DFID – The Bill & Melinda Gates Foundation – World Bank – Switzerland • Target areas: Marginal drought-prone environments – Africa (SSA) – South and South East Asia – Latin America • Mandate crops (CGIAR) – 22 crops • A CGIAR Challenge Programme hosted at CIMMYT, Mexico GCP Research Structure Director: Jean-Marcel Ribaut SP4 DB, Information Network (SPL: T van Hintum) Phenotyping Phenotyping SP1 Germplasm SP2 Gene Discovery SP3 MAS (SPL: J-C Glaszmann) (SPL: R K Varshney) (SPL: P Monneveux) SP5 Training/Capacity (SPL: C de Vicente) Breeding programmes (Delivery plans) Improved genotypes Improved germplasm in farmers’ fields Tropical Legume I project (GCP) US $ 11 m Objectives 1-4 Activity 1: Explore Diversity Objective 5 + Activity 2: Generate Genomic Resources Activity 3: Activity 4: Activity 5: Identify Marker Identify Marker Improve Development Development Germplasm (Biotic) (Abiotic) Development Objective 6 Peanut Cowpea Common bean Chickpea Comparative Genomics Training Objective 1- Peanut (PI: Dave Hoisington) Project Activities • Develop germplasm for genetic studies and modern breeding – Bonny Ntare (ICRISAT, Mali) • Generate genomic resources for genetic studies and modern breeding – Andy Paterson (University of Georgia, USA) • Identify molecular markers and genes for biotic stress resistance – David Bertioli (Catholic University, Brazil) • Identify molecular markers and genes for drought tolerance – Vincent Vadez (ICRISAT, India) • Improve locally adapted germplasm for target traits through modern breeding – Emmanuel Monyo (ICRISAT, Malawi) Objective 5- Cross species (PI: Doug Cook) Objective 5: Develop cross species resources for comparative genomics in tropical crop legumes Activity 1: Comparative Marker Development Milestones: Activity 2: Milestones: Activity 3: Milestones: Doug Cook 1. Development of molecular marker database and web interface. 2. Development of 500 orthologous gene markers. 3. Associate genetic markers to bacterial artificial chromosome clones in each species Analysis of the Arachis-species Complex David Bertioli 4. Diploid Arachis AA bacterial artificial chromosome clone end sequencing and SSR discovery 5. Mapping of orthologous genetic markers in diploid Arachis AA population 6. Identification of ultra-long SSRs in tetraploid Arachis 7. Genetic mapping of ultra-long SSRs in tetraploid Arachis Estimating Genome Divergence at Orthologous Loci Andy Paterson 8. DNA sequencing and divergence estimates for network of orthologous bacterial artificial chromosome clones Project partners • Catholic University of Brasilia, Brazil • Chitedze Research Station, Department of Research and Development, Ministry of Agriculture, Malawi • EMBRAPA, Brazil • ICRISAT, India/Malawi/Mali • Insitut National de Recherche Agronomique du Niger (INRAN), Niger • Institut Sénégalais de Recherche Agricole (ISRA), Senegal • Naliendele Research Station, Department of Research and Development, Ministry of Agriculture, Tanzania • University of Georgia, USA Take home message Community efforts Rapid advances in genomics Genome infrastructure available Applications to breeding Contributions and thanks Genomics: Rajeev Varshney Dave Hoisington Gautami, Ravi, Bryan, SomaRaju Genetic resources: Hari Upadhyaya Ranjana Bhattacherjee Breeding: Shyam Nigam, R Aruna Crop Physiology: Vincent Vadez Statistics & Bioinformatics: Subhash Chandra, Jayashree Collaboartions in India: NRC for Groundnut: T Radhakrishnan UAS Dharwad: MVC Gowda, Khedikar OVERSEAS COLLABORATIONS UC-Davis: Doug Cook, R Varma Catholic Univ: David Bertioli Tuskegee Univ: Guaho He Univ of Georgia: Steve Knapp Financial support: National Fund, Indian Council of Agricultural Research Generation Challenge Programme Department of Biotechnology



![9_Komlenac - start [kondor.etf.rs]](http://s2.studylib.net/store/data/005352037_1-bdc91b0717c49a75493200bca431c59c-300x300.png)