Modeling NMR Chemical Shifts
advertisement

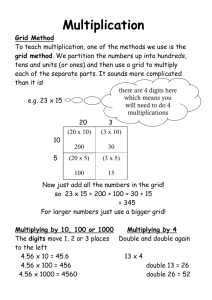
Gaussian 03 Calculations using GridNexus Ned H. Martin Department of Chemistry and Biochemistry University of North Carolina at Wilmington Grid Computing Rationale for Grid Computing The recent proliferation* of fast, interconnected underutilized cpus 14000 12000 10000 8000 MIPS 6000 ts/104 Series2 4000 2000 0 1960 1970 1980 1990 2000 2010 * over 150,000,000 pcs are sold each year! Grid Computing A computing Grid is analogous to an electrical power grid. The user simply “taps” into the resource (with permission), but is usually unaware of the origin of the resource. Grid Computing at UNCW Current efforts by a group of UNCW computer science faculty and undergraduate students, plus faculty and students in several “application areas” are focused on developing a graphical user interface (GUI) called GridNexus as a front-end to simplify data manipulations, searching or calculations of various types performed on remote computers over a Grid. This project has grant support from the UNC Office of the President GridNexus GridNexus is based on JXPL, a new graphical programming language developed by UNCW computer science faculty and students. GridNexus allows users to link modules that perform various operations into a usable ‘workflow’, then save these for later use. Once a ‘workflow’ has been created, one only need to specify the path/filename of the data set to be operated on and the path/filename for the output file. This greatly simplifies repetitive operations, and takes much of the mystery out of computing for non-computer science users. File Interconversion in GridNexus One of the limitations of most computational chemistry software packages is that they do not read or write many different (proprietary) file types, so it is difficult to transfer data from one program to another. GridNexus allows users to input the most common types of geometry specification, such as .pdb (.ent) and .mol files, and use a default set of options (or select from a list) to write a Gaussian input file (.dat). The structure can be oriented in XYZ coordinates as desired. Gaussian 03 under GridNexus Molecule Orientation in GridNexus One module allows a molecule to be oriented in Cartesian space in a specified way, then writes a proper Gaussian03 input file. Gaussian 03 Input File %chk=tmp/martinn/phenanthreneNH2.chk # HF/6-31G(d,p) opt freq Note C & N along the Y axis, the midpoint of their bond at the origin phenanthreneNH2 0 1 H C H C C N C C C C H H C (etc.) -1.963715 -1.127512 -0.184242 -0.149560 0.000000 0.000000 0.908090 -1.036579 0.971979 1.943981 -1.800364 1.238823 2.993024 -3.198017 -2.730904 -4.593909 -3.501921 -0.715690 0.715690 -2.892498 -1.338948 -1.491079 -3.742718 -0.744862 1.070292 -3.223318 1.280991 0.750482 0.244859 0.166986 0.000000 0.000000 -0.536779 0.691052 -0.702775 -1.057698 1.210005 -1.769705 -1.730309 Gaussian 03 under GridNexus Submitting a Gaussian job can be as simple as selecting the input file name (from a variety of file types) and the desired output file name. What’s next for GridNexus? Develop more “filters” to transform data. Enhance the graphics for appearance and usability. Include more software applications. Extend Grid services to other disciplines. Include industry and businesses as users and developers. Add more computational nodes to the Grid. The goal is to include all NC institutions of higher learning Acknowledgements • UNC-Office of the President • UNCW College of Arts and Sciences • UNCW Division of Academic Affairs •UNCW Department of Chemistry and Biochemistry • UNCW Information Technology Systems Division (ITSD) • Microelectronics Center of North Carolina (MCNC)