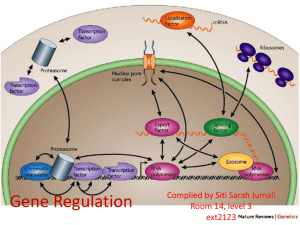
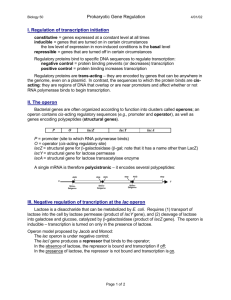
Lac operon Regulation of Transcription Initiation in Bacteria The
advertisement

Li Xiaoling Office: QQ: M1623 313320773 E-MAIL: 313320773 @qq.com 2016/3/21 Content Chapter 1 Introduction Chapter 2 The Structures of DNA and RNA Chapter 3 DNA Replication Chapter 4 DNA Mutation and Repair Chapter 5 RNA Transcription Chapter 6 RNA Splicing Chapter 7 Translation Chapter 8 The Genetic code Chapter 9 Regulation in prokaryotes Chapter 10 Regulation in Eukaryotes 2016/3/21 HOW TO LEARN THIS COURSE WELL? To preview and review Problem-base learning Making use of class time effectively Active participation Bi-directional question in class Group discussion Concept map Tutorship To call for reading, thingking and discussing of investigative learning 2016/3/21 To learn effectively EVALUATION (GRADING) SYSTEM in-class and attendance : 10 points Group study and attendance: 20 points Final exam: 70 points Bonus 2016/3/21 Question Molecular Biology of the Gene, 5/E --- Watson et al. (2004) Part I: Chemistry and Genetics Part II: Maintenance of the Genome Part III: Expression of the Genome Part IV: Regulation EXPRESSION OF THE GENOME Ch 5 : Transcription Ch 6 : RNA Splicing Ch 7 : Translation Ch 8 : The Genetic code Molecular Biology of the Gene, 5/E --- Watson et al. (2004) Part I: Chemistry and Genetics Part II: Maintenance of the Genome Part III: Expression of the Genome Part IV: Regulation Part V: Methods 7 REGULATION Ch 9: Regulation in prokaryotes Ch 10: Regulation in eukaryotes 8 Expression of many genes in cells are regulated Housekeeping genes: expressed constitutively, essential for basic processes involving in cell replication and growth. Inducible genes: expressed only when they are activated by inducers or cellular factors. 9 Chapter 9 Regulation principles and How genes are regulated in bacteria Chapter 10 Basic mechanism of gene expression in eukaryotes 10 Surfing the contents of Part IV --The heart of the frontier biological disciplines 11 Some of the peoples who significantly contribute to the knowledge of gene regulation 12 13 14 •Molecular Biology Course Chapter 9 Gene Regulation in Prokaryotes 15 TOPIC 1 Principles of Transcriptional Regulation [watch the animation] TOPIC 2 Regulation of Transcription Initiation: Examples from Bacteria (Lac operon, alternative s factors, NtrC,MerR, Gal rep, araBAD operon) TOPIC 3 Examples of Gene Regulation after Transcription Initiation (Trp operon) TOPIC 4 The Case of Phage λ: Layers of Regulation 16 CHAPTER 9 Gene Regulation in Prokaryotes Topic 1: Principles of Transcription Regulation 17 Principles of Transcription Regulation 1. GENE EXPRESSION IS CONTROLLED BY REGULATORY PROTEINS (调控蛋白) Gene expression is very often controlled by Extracellular Signals, which are communicated to genes by regulatory proteins: Positive regulators or activators INCREASE the transcription Negative regulators or repressors DECREASE or ELIMINATE the transcription 18 Principles of Transcription Regulation 2. GENE EXPRESSION IS CONTROLLED AT DIFFERENT STAGES (基因表达可以发生在不同时期) The bulk of gene regulation takes place at the initiation of transcription. Some involve transcriptional elongation/termination, RNA processing, and translation of the mRNA into protein. 19 FIG 9-3INITIATION Promoter Binding (closed complex) Promoter “melting” (open complex) Promoter escape/Initial transcription 20 FIG 9-3-ELONGATION AND TERMINATION Elongation Termination 21 Principles of Transcription Regulation 3. TARGETING PROMOTER BINDING: MANY PROMOTERS ARE REGULATED BY ACTIVATORS (激活蛋白) THAT HELP RNAP BIND DNA (RECRUITMENT) AND BY REPRESSORS (阻遏蛋白) THAT BLOCK THE BINDING. RNAP binds many promoters weakly (?), activators that contain two binding sites to bind a DNA sequence and RNAP simultaneously can enhance the RNAP affinity with the promoters, and thus increases gene transcription.This is called recruitment regulation (招募调控). On the contrary, Repressors can bind to the operator inside of the promoter region, which prevents RNAP binding and the transcription 22 of the target gene. a. Absence of Regulatory Proteins: basal level expression Fig 9-1 b. Repressor binding to the operator represses expression c. Activator binding activates expression 23 Principles of Transcription Regulation 4 Targeting transition to the open complex: Allostery regulation (异构调控) after the RNA Polymerase Binding In some cases, RNAP binds the promoters efficiently, but no spontaneous isomerization occurs to lead to the open complex, resulting in no or low transcription. Some activators can bind to the closed complex, inducing conformational change in either RNAP or DNA promoter, which converts the closed complex to open complex and thus promotes the transcription. 24 Allostery regulation Fig 9-2 Allostery is not only a mechanism of gene activation , it is also often the way that regulators are controlled by their specific signals. 25 Principles of Transcription Regulation 5 Targeting promoter escape by some repressors Repressors can work in ways: (1) blocking the promoter binding. (2) blocking the transition to the open complex. (3) blocking promoter escape 26 Some promoters are inefficient at more than one step and can be activated by more than one mechanism. Activation mechanisms include recruitment (招募) and allostery (异构). 27 Principles of Transcription Regulation 6. Cooperative binding (recruitment) and allostery have many roles in gene regulation For example: group of regulators often bind DNA cooperatively (activators and/or repressors interact with each other and with the DNA, helping each other to bind near a gene they regulated) : (1) produce sensitive switches to rapidly turn on a gene expression, (2) integrate signals (some genes are activated when multiple signals are present). 28 Principles of Transcription Regulation 7. Action at a Distance and DNA Looping. The regulator proteins can function even binding at a DNA site far away from the promoter region, through protein-protein interaction and DNA looping. Fig 9-3 29 Fig 9-4 DNA-binding protein can facilitate interaction between DNAbinding proteins at a distance Fig 9-4 30 CHAPTER 9 Gene Regulation in Prokaryotes Topic 2: Regulation of Transcription Initiation : Examples from Bacteria 31 Operon: a unit of prokarytoic gene expression and regulation which typically includes: 1. Structural genes for enzymes in a specific biosynthetic pathway whose expression is coordinately controlled. 2. Control elements, such as operator sequence. 3. Regulator gene(s) whose products recognize the control elements. 32 Sometimes are encoded by the gene under the control of a different promoter Control element Structural genes 33 Regulation of Transcription Initiation in Bacteria First example: Lac operon The lactose Operon 34 1. Lactose operon contains a regulatory gene and 3 structural genes, and 2 control elements. Fig 9-5 The enzymes encoded by lacZ, lacY, lacA are required for the use of lactose as a carbon source. These genes are only transcribed at a high level when lactose is 35 available as the sole carbon source. The LAC operon lacZ codes for β-galactosidase (半乳 糖苷酶) for lactose hydrolysis lacY encodes a cell membrane protein called lactose permease (半乳糖苷渗透酶) to transport Lactose across the cell wall lacA encodes a thiogalactoside transacetylase (硫代半乳糖苷转 乙酰酶)to get rid of the toxic 36 thiogalacosides The LAC operon THE LACZ, LACY, LACA GENES TRANSCRIBED INTO A ARE SINGLE LACZYA MRNA (POLYCISTRONIC MRNA) UNDER CONTROL OF A SIGNAL PROMOTER THE PLAC . LacZYA transcription unit contains an operator site Olac position between bases -5 and +21 at the 3’-end of Plac Binds with the lac repressor 37 The LAC operon 2. An activator and a repressor together control the Lac operon expression The activator: CAP (Catabolite Activator Protein,代谢产物激活蛋白) or CRP (cAMP Receptor Protein,cAMP受体蛋 白); responses to the glucose level. The repressor: lac repressor that is encoded by LacI gene; responses to the lactose. 38 Sugar switch-off mechanism The LAC operon The LAC operon Fig 9-6 39 The LAC operon 3. Lac repressor bound to the operator prevents RNAP from binding to the promoter The site bound by lac repressor is called the lac operator (Olac ), and the Olac overlaps promoter (Plac). Therefore repressor bound to the operator physically prevents RNA polymerase from binding to the promoter. 40 The LAC operon The LAC operon Fig 9-8 41 The LAC operon 4. CAP activates the Lac transcription through recruitment of RNAP to the weak Plac CAP has two binding sites, one interacts with the CAP site on the DNA near promoter, and one interacts with RNAP. This cooperative binding ensures that RNAP effectively binds to Plac and initiates transcription of LacZYA. 42 The LAC operon CAP site has the similar structure as the operator, which is 60 bp upstream of the start site of transcription. CAP also interacts with the RNAP and recruit it to the promoter. Fig 9-9 a CTD: C-terminal domain of the a subunit of RNAP 43 The LAC operon CAP binds as a dimer a CTD Fig 9-10. CAP has separate activating and DNA-binding surface 44 5. CAP and Lac repressor bind DNA using a common structural motif: helix-turn-helix motif Fig 9-11 One is the recognition helix that can fits into the major groove of the DNA. The LAC operon 45 DNA binding by a helix-turn-helix motif Fig 9-12 Hydrogen Bonds between l repressor and the major groove of the operator. 46 Lac operon contains three operators: the primary operator and two other operators located 400 bp downstream and 90 bp upstream. Lac repressor binds as a tetramer (四聚体), with each operator is contacted by a repressor dimer (二聚体). respectively. Fig 9-13 47 6 The activity of Lac repressor and CAP are controlled allosterically by their signals. Allolactose: turn of Lac repressor cAMP: turn on CAP Lactose is converted to allolactose by bgalactosidase, therefore lactose can indirectly turn off the repressor. Glucose lowers the cellular cAMP level, therefore, glucose indirectly turn off CAP. 48 The LAC operon Response to lactose Lack of inducer: the lac repressor block all but a very low level of transcription of lacZYA . Absence of lactose i p o z y a Active Very low level of lac mRNA When Lactose is present, the low basal level of permease allows its uptake, and b-galactosidase catalyzes the conversion of some lactose to allolactose. Allolactose acts as an inducer, binding to the lac repressor and inactivate it. Presence of lactose i p o z y a Inactive Permease Transacetylase b-Galactosidase 49 Response to glucose 50 7: Combinatorial Control (组合调 控): CAP controls other genes as well A regulator (CAP) works together with different repressor at different genes, this is an example of Combinatorial Control. In fact, CAP acts at more than 100 genes in E.coli, working with an array of partners. 51 Regulation of Transcription Initiation in Bacteria Second example: Alternative s factor Alternative s factor (可变s因子) direct RNA polymerase to alternative site of promoters 52 s factor subunit bound to RNA polymerase for transcription initiation (Ch 12) 53 Different s factors binding to the same RNAP, conferring each of them a new promoter specificity. s70 factors is most common one in E. coli under the normal growth condition 54 Many bacteria produce alternative sets of σfactors to meet the regulation requirements of transcription under normal and extreme growth condition. Bacteriophage has its own σfactors E. coli : Heat shock s32 Bacteriophage σfactors Sporulation in Bacillus subtilis 55 HEAT SHOCK (热休克) Around 17 proteins are specifically expressed in E. coli when the temperature is increased above 37ºC. These proteins are expressed through transcription by RNA polymerase using an alternative s factor s32 coded by rhoH gene. s32 has its own specific promoter consensus sequences. 56 Alternative s factors Bacteriophages Many bacteriophages synthesize their own σfactors to endow the host RNA polymerase with a different promoter specificity and hence to selectively express their own phage genes . 57 Alternative s factors Alternative s factors Fig 9-14 B. subtilis SPO1 phage expresses a cascade of σfactors which allow a defined sequence of expression of different phage genes. 58 Regulation of Transcription Initiation in Bacteria Third example: NtrC and MerR and allosteric activation Transcriptional activators NtrC and MerR work by allostery rather than by recruitment. 59 Review The majority of activators work by recruitment, such as CAP. These activators simply bring an active form of RNA polymerase to the promoter In the case of allosteric activation, RNAP initially binds the promoter in an inactive complex, and the activator triggers an allosteric change in that complex to activate transcription. 60 In the absence of NtrC and MerR, RNAP binds to the corresponding promoter to form a closed stable complex. NtrC activator induces a conformational change in the enzyme, triggering transition to the open complex MerR activator causes the allosteric effect on the DNA and triggers the transition to the open complex 61 NtrC and MerR and allosteric activation 1. NtrC has ATPase activity and works from DNA sites far from the gene NtrC controls expression of genes involved in nitrogen metabolism (氮代谢), such as the glnA gene NtrC has separate activating and DNA-binding domains, and binds DNA only when the nitrogen levels are low. 62 Low nitrogen levels (低水平氮)NtrC phosphorylation and conformational change NtrC (?) binds DNA sites at ~-150 positio as a dimer NtrC (?) interacts with s54 (glnA promoter recognition) NtrC ATPase activity provides energy needed to induce a conformation change in polymerase transcription STARTs Fig 9-15 activation by NtrC 63 NtrC and MerR and allosteric activation 2. MerR activates transcription by twisting promoter DNA MerR controls a gene called merT, which encodes an enzyme that makes cells resistant to the toxic effects of mercury (抗汞酶) In the presence of mercury (汞), MerR binds to a sequence between –10 and –35 regions of the merT promoter and activates merT expression. 64 As a s70 promoter, merT contains 19 bp between –10 and –35 elements (the typical length is 15-17 bp), leaving these two elements recognized by s70 neither optimally separated nor aligned. 65 66 Fig 9-15 Structure of a merT-like promoter When Hg2+ is absent, MerR binds to the promoter and locks it in the unfavorable conformation When Hg2+ is present, MerR binds Hg2+ and undergoes conformational change, which twists the promoter to restore it to the structure close to a strong s70 promoter Fig 9-15 67 Repressors work in many ways-review Blocking RNA polymerase binding through binding to a site overlapping the promoter. Lac repressor Blocking the transition from the closed to open complex. Repressors bind to sites beside a promoter, interact with polymerase bound at that promoter and inhibit initiation. E.coli Gal repressor Blocking the promoter escape. P4 protein interaction with PA2c (bacteriophage f29 ) 68 Regulation of Transcription Initiation in Bacteria Fourth example: araBAD operon 69 The araBAD operon 1. AraC and control of the araBAD operon by anti-activation The promoter of the araBAD operon from E. coli is activated in the presence of arabinose (阿拉伯 糖) and the absence of glucose and directs expression of genes encoding enzymes required for arabinose metabolism. This is very similar to the Lac operon. 70 Different from the Lac operon, two activators AraC and CAP work together to activate the araBAD operon expression 194 bp CAP site Fig 9-18 71 Because the magnitude of induction of the araBAD promoter by arabinose is very large, the promoter is often used in expression vector. If fusing a gene to the araBAD promoter, the expression of the gene can be easily controlled by addition of arabinose(阿拉伯糖). What is an expression vector ? [The answer is in the Methods part.] 72 CHAPTER 9 Gene Regulation in Prokaryotes Topic 3: Examples of Gene Regulation at Steps After Transcription Initiation 73 Examples of Gene Regulation at Steps After Transcription Initiation First example: the tryptophan operon (色 胺酸操纵子) 74 1. Amino acid biosynthetic operons are controlled by premature transcription termination: the trp operon 75 The TRP operon The trp operon encodes five structural genes required for tryptophan (色胺酸) synthesis. These genes are regulated to efficiently express only when tryptophan is limiting. Two layers of regulation are involved: (1) transcription repression by the Trp repressor (initiation); (2) attenuation 76 The TRP operon The Trp repressor (色氨酸阻遏物 ) 77 The TRP operon 1. 2. Trp repressor is encoded by a separate operon trpR, and specifically interacts with the operator that overlaps with the promoter sequence The repressor can only bind to the operator when it is complexed with tryptophan. Therefore, Try is a corepressor and inhibits its own synthesis through end-product inhibition (negative feed-back regulation). Remember the lac repressor acts as an inducer 78 The TRP operon 3. The repressor reduces transcription initiation by around 70-fold, which is much smaller than the binding of lac repressor. 4. The repressor is a dimer of two subunits which has a structure with a central core and two flexible DNA-reading heads (carboxyl-terminal of each subunit ) 79 The TRP operon trpR operon trp operon 80 The TRP operon Attenuation (衰减作用) : a regulation at the transcription termination step & a second mechanism to confirm that little tryptophan is available 81 Repressor serves as the primary switch to regulate the expression of genes in the trp operon Attenuation serves as the fine switch to determine if the genes need to be efficiently expressed 82 Fig 9-19 Transcription of the trp operon is prematurally stopped if the tryptophan level is not low enough, which results in the production of a leader RNA of 161 nt. (WHY?) 83 1. 2. 3. Transcription and translation in bacteria are coupled (细菌体内的转录和翻译是偶联的). Therefore, synthesis of the leader peptide immediately follows the transcription of leader RNA. The leader peptide contains two tryptophan codons. If the tryptophan level is very low, the ribosome will pause at these sites. Ribosome pause at these sites alter the secondary structure of the leader RNA, which eliminates the intrinsic terminator structure and allow the successful 84 transcription of the trp operon. 85 Fig 9-20 The leader RNA and leader peptide High Trp Complementary 3:4 termination of transcription Low Trp Complementary 2:3 Elongation of transcription Fig 9-21 86 Importance of attenuation 1. 2. 3. 4. A typical negative feed-back regulation Use of both repression and attenuation allows a fine tuning of the level of the intracellular tryptophan. Attenuation alone can provide robust regulation: other amino acids operons like his and leu have no repressors and rely entirely on attenuation for their regulation. Provides an example of regulation without the use of a regulatory protein, 87 but using RNA structure instead. Examples of Gene Regulation at Steps After Transcription Initiation Second example: Riboswitches-a RNA structure control mechanism Riboswitches are regulatory RNA elements that act as direct sensors of small molecule metabolites to control gene transcription or translation. 88 Box 4 1.Riboswitches operating at the level of transcription termination using an Antitermination mechanism. 2.Riboswitches operating at the level of translation, controlling the formation of an RNA structure that masks the ribosome binding site on mRNA. 89 代谢物 代谢物 90 Tucker1 and Breaker, Current Opinion in Structural Biology 2005, 15:342–3 The 2nd structures of 7 riboswitches and metabolites that they sense 91 Examples of Gene Regulation at Steps After Transcription Initiation Third example: Ribosomal proteins are translational repressors of their own synthesis: a negative feedback 92 Challenges the ribosome protein synthesis 1. Each ribosome contains some 50 distinct proteins that must be made at the same rate. 2. The rate of the ribosome protein synthesis is tightly closed to the cell’s growth rate. 93 Strategies to meet the challenges-Operon 1. Organization of the ribosomal proteins to several operons (操纵子) , each containing up to 11 ribosomal protein genes 2. Some nonribosomal proteins whose synthesis is also linked to growth rate are contained in these operons, including those for RNAP subunits a, b and b’. 3. The primary control (主要调控) is at the level of translation, not transcription.94 Ribosomal protein operons The protein that acts as a translational repressor of the other proteins is shaded red. Fig 9-22 95 Strategies to meet the challenges (cont): 4. 5. 6. For each operon, one (or two) ribosomal proteins binds the mRNA near the translation initiation sequence, preventing the ribosome from binding and initiating translation. Repressing translation of the first gene also prevents expression of some or all of the rest. The strategy is very sensitive. A few unused molecule of protein L4, for example, will shut down synthesis of that protein and 96 other proteins in this operon. 7. The mechanism of one ribosomal protein also functions as a regulator of its own translation: the protein binds to the similar sites on the ribosomal RNA and to the regulatory RNA in its own mRNA. Fig 9-23 97 Key points of the chapter 1. Principles of gene regulation. (1) The targeted gene expression events; (2) the mechanisms: by recruitment/exclusion or allostery 2. Regulation of transcription initiation in bacteria: the lac operon, alternative s factors, NtrC, MerR, Gal rep, araBAD operon 3. Examples of gene regulation after transcription initiation: the trp operon, riboswitch, regulation of the synthesis of ribosomal proteins 98