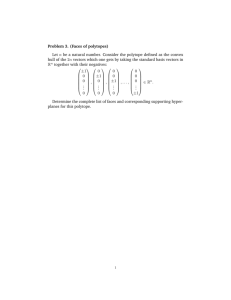
(Right) Null Space of S Systems Biology by Bernhard
advertisement

The (Right) Null Space of S Systems Biology by Bernhard O. Polson Chapter9 Deborah Sills Walker Lab Group meeting April 12, 2007 Overview • Stoichiometric matrix • Null Space of S • Linear and Convex basis vectors for the null space • Extreme Pathway Analysis • Practical applications S connectivity matrix– represents a network A v1 b1 b2 1 S 1 1 1 0 0 0 1 0 b1 b3 0 0 1 B A B C v1 b2 C Each column represents reaction (n reactions) Each row represents a metabolite (m metabolites) b3 Stoicheometric Matrix • S is a linear transformation of of the flux vector dx Sv dt x = metabolite concentrations (m, 1) S = stoicheometric matrix (m,n) v = flux vector (n,1) Four fundamental subspaces • Column space, left null space, row space, and (right) null space Iman Famili and Bernhard O. Palsson, Biophys J. 2003 July; 85(1): 16–26. Null Space of S • Steady state flux distributions (no change in time) dx Sv 0 dt Svs s 0 Basis of the Null Space • Basis spans space of matrix • Null space spanned by (n-r) basis vectors Where n = number of metabolites r = rank of S (number of linearly independent rows and columns) • Exampes of bases: linear basis, orthonormal basis, convex basis Basis of Null Space contin. •Null space orthogonal to row space of S •Basis vectors form columns of matrix R ri i[1, n-r] SR= 0 •Every point in vector space, uniquely defined by set linearly independent basis vectors v s s wi ri Choosing a Basis for a Biological Network Two types of reactions in open systems: – Elementary reactions (internal) only have positive flux v i 0 – Exchange fluxes can include diffusion and are considered bidirectional 0 bi 0 A b1 B v1 b2 C b3 B Null Space defined by: v3 v2 v1 r1 A v4 r2 v5 D v6 1 0 0 0 v1 1 0 0 1 0 v2 0 1 1 0 0 0 v3 0 0 1 1 0 1 v4 0 0 0 0 1 1 v5 0 v 6 C Matrix full rank, and dimension of null space r-n = 6 - 4 = 2 Column 4, 6 don’t contain pivot, so solve null space in terms of v4 and v6 1 0 1 1 1 1 w1r1 w2r2 v v4 v6 1 0 0 1 0 1 Nonnegative linear basis vectors for null space r1, r2 1 1 1 1 0 0 0 1 1 0 1 1 1 1 0 1 1 1 1 1 0 0 1 0 0 p1 , p2 1 1 1 p1 B 1 1 1 p1 , p2 1 0 0 1 0 0 1 1 1 v3 v2 A v4 v1 D v5 v6 C p2 Biologically irrelevant since no carrier or cofactor (such as ATP) exchanges Convex Bases • Convex bases unique • Number of convex basis bigger than the dimension of the null space • Elementary reactions positive and have upper bound 0 vi vi max Allowable fluxes are in a hyperbox bounded by planes parallel to each axis as defined by vi,max Convex Basis •Hyperbox contains all fluxes (steady state and dynamic) •Sv=0 is hyperplane that intersects hyperbox forming finite segment of hyperplane •Intersection is polytope in which all steady state fluxes lie •Polytope spanned by convex basis vectors, which are edges of polytope, with restricted ranges on weights v ss k p k k 1 Convex basis vectors are edges of polytope that contain steady state flux vectors v ss k p k k 1 Where pk are the edges, or extreme states, and k are the weights that are positive and bounded, 0 i ,min i i ,max Aside(from Schilling et al., BiotechBioeng.2000. •Convex polyhedron is a region in Rn determined by linear equalities and inequalities •Polytope is bounded polyhedron •Polyhedral cone if every ray through the origin and any point in the polyhedron are completely contained in polyhedron Simple 3-D example •Node with three reactions forms simple flux split •Min and max constraints form box that is intercepted by plain formed by flux balance 0=v3 – v1- v2= [(-1, -1, 1)•(v1, v2 , v3)] •2D polytope spanned by two convex basis vectors b1= (v1, 0, v3) b2= (0, v2, v3) Null space links biology and math • Null space represents all functional, phenotypic states of network • Each point in polytope represent one network function or one phenotypic state • Edges of flux cone are unique extreme pathways • Extreme pathways describe range of fluxes that are permitted Constraints in Biological Systems • • • • • • • Thermodynamic – reversibility Mass balance Maximum enzyme capacity Energy balance Cell volume Kinetics Transcriptional regulatory constraints Constraints in Biological Systems contin. • Constraints can help to determine effect of various parameters on achievable states of network • Examples: enzymopathies can reduce certain maximum fluxes, reducing i,max • Effects of gene deletion can also be examined Biochemical Reaction Network and its convex, steady-state solution cone v ss k p k k 1 0 k k ,max [Nathan D. Price, Jennifer L. Reed, Jason A. Papin, Iman Famili and Bernhard O. Palsson , Analysis of Metabolic Capabilities Using Singular Value Decomposition of Extreme Pathway Matrices. The Biophysical Society, 2003.] Classification of Extreme Pathways p1……………………………………………pk v1 Type I . Primary system . 0 . . vn b1 0 bnE Type II Futile cycle Type III Internal cycles 0 0 0 0 Internal fluxes Exchange fluxes Extreme Pathways • Type I involve conversion of primary inputs to primary outputs • Type II involve internal exchange carrier metabolites such as ATP and NADH • Type III are internal cycles with no flux across system boundaries Extreme Pathways Extreme Skeleton Metabolic Pathways • Glycolysis has five extreme pathways – Two type I represent in secretion of two metabolites – One type II represent dissipation of phosphate bond – futile pathway – Two type III that will have no net flux Basis Vectors for Biological System • ri makes the nodes in the flux map “link neutral”, because it is orthogonal to all the rows of S simultaneously • Network-based pathway definition, and use basis vectors (pi) that represent these pathways Practical Applications • Convex bases offers a mathematical analysis of the null space that makes biochemical sense • Flux-balance analysis mostly used so far to analyze single species • Analysis of complex communities challenge, but possible to limit study to core model • Stolyar et al., 2007 used multispecies stoicheometric metabolic model to predict mutualistic interactions between sulfate reducing bacteria and methanogen