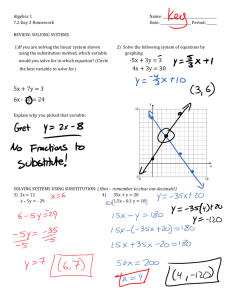
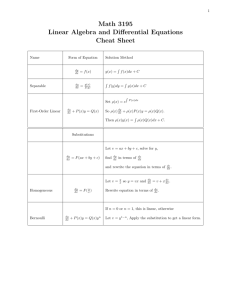
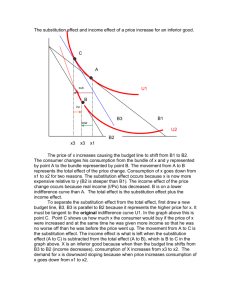
Document
advertisement

Measuring genetic change Level 3 Molecular Evolution and Bioinformatics Jim Provan Page and Holmes: Section 5.2 Types of substitution A C A A T C C A T C G A C G A C A A A Single 1 change, 1 difference Multiple 2 changes, 1 difference Coincidental 2 changes, 1 difference C C A C A C T C T A T C A T A A A C C A A A A Parallel 2 changes, no difference Convergent 3 changes, no difference Back 2 changes, no difference Types of substitution (continued) Multiple substitutions can greatly obscure actual evolutionary history, particularly in cases where there have been many mutations i.e. over long evolutionary time scales Final three examples have serious implications for inference of evolutionary history: Similarity inherited from an ancestor is called homology Independently acquired similarity is called homoplasy All tree-building methods rely on sufficient levels of homology Types of substitution (continued) A C G T Substitutions that exchange a purine for another purine or a pyrimidine for another pyrimidine are called transitions Substitutions that exchange a purine for a pyrimidine or vice-versa are called transversions Measuring evolutionary change Some sites may undergo repeated substitutions As sequences diverge, measure becomes less accurate Saturation occurs most sites changing have changed before 120 Base pair differences Simplest measure is to count number of different sites Poor measure: 100 80 60 40 20 0 0 5 10 15 20 Time since divergence (Myr) 25 Correction of observed sequence differences Sequence difference Expected difference ‘Correction’ Observed difference Time A general framework of sequence evolution models pAA Pt = pAC pAG pAT pCA pCC pCG pCT pGA pGC pGG pGT pTA pTC pTG pTT Pii = 1 - ji f = [fA fC fG fT] pij The Jukes-Cantor (JC) model Assumes that all four bases have equal frequencies and that all substitutions are equally likely Pt = - - - - f = [¼ ¼ ¼ ¼] Kimura’s 2 parameter model (K2P) Takes into account different frequencies of transitions vs. transversions Pt = 100 90 80 70 - - - - Transitions () 60 50 40 30 Transversions () 20 f = [¼ ¼ ¼ ¼] 10 0 0 5 10 15 20 25 Felsenstein (1981) (F81) Takes into account differences in base composition Percentage (G + C) can range from 25% - 75% F81 model allows the frequencies of the four nucleotides to be different Does not allow for variation between genes/species C G T Pt = A - A C G T - A C G T - f = [A C G T] Hasegawa, Kishino and Yano (1985) (HKY85) Essentially merges the K2P and F81 models to allow transitions and transversions to occur at different rates as well as allowing base frequencies to vary Pt = A C G T - A C G T - A C G T - f = [A C G T] General reversible model (REV) Most general model - each substitution has its own probability Pt = Aa Ca Gb - Ab Cd Ac Ce Tc Gd Te - Tf Gf - f = [A C G T] By constraining a-f it is possible to generate all the other models Comparing the models Allow transition/ transversion bias JC K2P A=C=G=T Allow base frequencies to vary HKY85 REV A=C=G=T ACGT ACGT = a,b,c,d,e,f Allow base frequencies to vary F81 ACGT = Allow transition/ transversion bias Comparing the models (continued) A Observed C G T A A C C K2P G T C G T A C C T G T A C G T G A G C T A JC A HKY85 G T Assumptions: independence Assumes that change at one site has no effect on other sites Good example is in RNA stem-loop structures Substitution may result in mismatched bases and decreased stem stability Compensatory change may occur to restore Watson-Crick base pairing A G A C C C CU U GGGG A A G C A U G C C C C U U C A GGG C A A G U A G C C CGU U GGG C A A G C A U Assumptions: base composition Assumption that base composition is at equilibrium and that it is similar across all taxa studied In example opposite, trees inferred using models which do not allow for this will not group Thermus and Deinococcus %G+C Aquifex 64.0 Thermotoga 63.7 Thermus 63.2 Deinococcus 55.5 Others 53.9 All sites are not equally likely to undergo a substitution Functional constraints: Pseudogenes have lost all function and can evolve freely Fourfold degenerate sites do not change amino acid composition of proteins Non-degenerate sites are highly constrained Substitution / site / 109 years Assumptions: variation in substitution rate across sites 4 3.5 3 2.5 2 1.5 1 0.5 0 Assumptions: variation in substitution rate across sites (continued) 0.7 DNA divergence 0.5% / Myr + 20% constraint A 0.6 0.5 0.4 2% / Myr + 50% constraint B 0.3 0.2 0.1 0 0 50 100 150 200 250 Divergence time (Myr) More rapidly evolving sequence shows most divergence initially but soon saturates Sequence A actually appears to be more rapidly evolving