File
advertisement

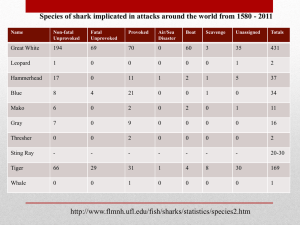
Bio-Identification of Flake Samples Using DNA Barcoding By Lina Martinez, Jeremy Tallman, Richard Moloney, Chantall Smith, Joanna Wisniewski , Wathsala Kumari Katherine Te DNA BARCODING •Provides a faster method of accuracy determining a species when compared with morphological identification. •It allows for how species are assembled within a biological systems . •Determine the composition/purity of biological samples . •The DNA Barcode used here is a section of the Cytochrome c oxidase 1, a region involved in the electron phase of mitochondrial respiration. Our group decided to determine if DNA Barcoding could be used to identify which species of shark are sold in Australia as “flake”. Hypothesis – that different species of shark are sold under the banner of flake Primer sequences used in the experiment were: “A fragment from the 5’ end of COI (~ 648 base pairs) is isolated (the entire gene has~ 1500 base pairs).” COI barcoding (fish) 5’-TGTAAAACGACGGCCAGTCAACCAACCACAAAGACATTGGCAC -3’ α FORWARD PRIMER – VF2_t1 5’-TGTAAAACGACGGCCAGTCGACTAATCATAAAGATATCGGCAC-3’ α FORWARD PRIMER – Fish F2_t1 5’-CAGGAAACAGCTATGACACTTCAGGGTGACCGAAGAATCAGAA-3’ α REVERSE PRIMER – Fish R2_t1 5’-CAGGAAACAGCTATGACACCTCAGGGTGTCCGAARAAYCARAA-3’ α α α REVERSE PRIMER – FR1 For other materials and methods refer to Project Manual (Diploma In Laboratory Techniques MSL50109) We had DNA of of about 580 base pairs which was the expected size. CONSENSUS SEQUENCES FLAKE SAMPLE - Galeorhinus galeus TGGGACAGCCAGGATCTCTTTTAGGAGATGACCAGATTTATAATGTGATCGTAAC CGCCCATGCTTTTGTAATAATCTTTTTTATGGTTATACCAATCATGATTGGTGGCT TTGGAAATTGACTAGTTCCATTAATAATTGGTGCGCCTGATATAGCCTTCCCACG GATAAATAACATAAGCTTCTGACTTCTTCCACCATCATTCCTTCTTCTCCTAGCTT CTGCCGGAGTAGAAGCTGGAGCAGGTACTGGTTGAACAGTATATCCTCCACTAG CAAGCAATTTAGCCCATGCTGGACCATCTGTAGATTTAGCCATTTTCTCCCTTCA TTTAGCCGGTATCTCATCAATCCTAGCCTCAATTAATTTTATTACAACCATTATTA ACATAAAACCCCCAGCTATTTCCCAATATCAAACACCATTATTTGTTTGATCAATT CTTGTAACTACTATTCTTCTTCTCCTCTCTCTCCCAGTTCTCGCAGCAGGAATCAC AATATTACTTACAGACCGTAACCTTAATACCACATTCTTTGACCCTGCAGGTGGA GGAGACCCAATCCTTTACCAACATTTATTCTGATTCTTC SAMPLE NUMBER COLLECTION LOCATION NAME OF SPECIES Flake 1 Fish shop (fresh) Mustelus lenticulatus- Spotted Estuary Smooth-Hound Flake 2 Fish shop (cooked) Flake 3 Supermarket (fresh) Flake 4 Market (fresh) Mustelus antarcticus – Gummy Shark Galeorhinus galeus- School Shark, Tope Shark, Soupfin Shark or Snapper Shark Mustelus antarcticus – Gummy Shark Flake 5 Market (fresh) Flake 6 Market (fresh) Flake 7 Supermarket (fresh) Table 1. Results of DNA sequencing No Results No Results Galeorhinus galeus- School Shark, Tope Shark, Soupfin Shark or Snapper Shark “Just as a unique pattern of bars in a universal product code (UPC) identifies each item for sale in a store, a DNA barcode is a DNA sequence that uniquely identifies each species of living thing” 1 DNA barcoding has been successful in identifying the species of shark samples used in our experiment. 7 samples of Flake were purchased from different locations in Melbourne. Genomic DNA was extracted, and used for PCR. 2 of the 7 samples had a low quality result and were removed from the data set. 5 were sufficient enough for analysis. •Mustelus antarcticus, is a shark in the family triakidae . It is a slender, grey shark with white spots along the body and flat, plate-like teeth for crushing its prey. •It has a maximum length of between 157 cm (male) and 175 cm (female). It is found in the waters around southern Australia, from Shark Bay in WA to Port Stephens in NSW to a depth of 350 m. •Marketed as "flake”. GUMMY SHARK http://www.oceanwideimages.com/images/10801/large/gum my-shark-24M2642-01D.jpg • Mustelus lenticulatus, is a houndshark of the family triakidae, found on the continental shelves and in estuaries around southern Australia and New zealand . •Its length is up to 125 cm (male) and 151 cm (female). •“FISH AND CHIP” = FLAKE SPOTTED ESTUARY SMOOTH-HOUND http://www.southern-edge.com/rig-shark-or-lemonfish/ • Galeorhinus galeus, is a hound shark of the family Triakidae. • Found worldwide in subtropical seas at depths of up to 550 metres (1,800 ft). • It grows to 2 metres (6 ft 7 in) long. SCHOOL SHARK, TOPE SHARK, SOUPFIN SHARK OR SNAPPER SHARK, http://www.exploreuw.com/Galeorhinus_galeus.d Our results support our hypothesis. We found 3 different species are sold as “flake”. This shows that DNA Barcoding can be used to identify seafood species sold in Australia. 2 out of the 5 shark DNA sequences was that of the endangered Tope Shark. The Tope Shark is found on the Red List of the IUCN - The World Conservation Union. DNA barcoding can be used to help in the conservation of endangered species. The government has a National Plan of Action for the Conservation and Management of Sharks 2012 – Shark Plan 2 It aims on working on conservation and managing Australian Fisheries. Our data suggests that DNA barcoding could be a good method to assist this plan For future experiments a bigger sample size would help to discover how wide spread endangered species are being consumed and labelled as “shark”. The larger the sample size of shark being tested, the more we can have confirmation of our results. Our results agree with the results of the students from New York’s Trinity School who did an experiment on ‘mislabelled’ fish. We found out that DNA Barcoding is an effective technique and can be use to identify different species of sharks sold in the supermarket as “flake”.