Folie 1

Nature, 24 July 2008; 454(7203):523-527
05.01.2009, Wiebke Albrecht
Innate immunity and virus recognition
innate immunity is characterized by the use of pattern recognition receptors (PRRs)
PRRs recognize pathogenassociated molecular patterns
(PAMPS)
PRR activation leads to type I interferon (IFN) and proinflammatory cytokine production
3 types: toll-like receptors (TLRs)
RIG-I-like receptors (RLRs)
NOD-like receptors
RIG-I = retinoic acid-inducible gene I
NOD = nucleotide-binding oligomerization domain
PRRs induce type I IFN and proinflammatory cytokines
Saito et al. (2008), commentary
RIG-I
located in the cytoplasm
consists of a DEXD/H box helicase domain, two CARD-like domains required for activation of downstream signalling pathways and a C-terminal domain (CTD) that includes a repressor domain (RD) inhibiting signalling in the steady state
recognizes short dsRNA and 5‘ terminal triphosphate RNA (5‘ ppp RNA) binding of nonself RNA to RIG-I leads to a conformational change, the CARD domains are exposed and activate downstream signalling
CARD = caspase-recruiting domain
Model of RIG-I activation
Yoneyama et Fujita (2008)
Hepatitis C virus (HCV)
small enveloped virus (diameter ~50nm)
(+) ssRNA genome
genus: Hepacivirus family: Flaviviridae
causes chronic hepatitis, liver cirrhosis and hepatocellular carcinoma
six major genotypes are known, differing in their geographic distribution and their responsiveness to antiviral therapy
HCV life cycle
Lindenbach et al. (2005)
HCV genome
genome size: ~9,6 kb
genome encodes one ORF which is translated as a polyprotein and subsequently cleaved into ten proteins
5‘ NTR is a conserved region and consists of four domains, including an IRES element to direct cap-independent translation
3‘ NTR consists of 3 domains sufficient for replication
HCV polyprotein
Bode et al. (2008)
ORF = open reading frame
NTR = non-translated region
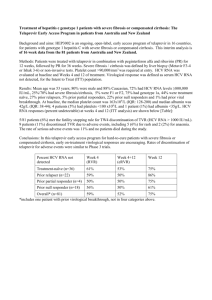
Identification of HCV PAMP RNA motifs
luciferase reporter assay with a reporter plasmid containing an IFNβ promotor
carried out in Huh7 cells (human hepatoma cell line) two regions inducing the IFNβ promotor, further mapping of the responsible regions
IFN = interferon
ORF 3‘ NTR
further mapping to nt 2406-
2696 of the ORF and to nt
93899616 of the 3‘ NTR
deletion of of the 3‘ NTR but not deletion of the region nt 2408-2663 attenuated promotor signalling
PAMP motifs are typically conserved, nt 9389-9616 show high conservation between different HCV starins nt = nucleotides
Structure of the HCV 3‘ NTR
VR = variable region with potential secondary structure (~40 nt)
PU/UC = non-structured poly (U/UC) tract containing polyuridine with interspersed ribocytidine (variable length)
X = highly conserved segment that formes 3 stemloop structures (~98 nt)
RI = replication intermediate (- RNA strand)
HCV PAMP in the viral 3‘ NTR
reporter assay in Huh7
PU/UC region is sufficient for signal triggering
also shown in Hela cells
IRF = interferon regulatory factor
ISG = antiviral/interferon-stimulated gene
full legth 3‘ NTR as well as PU/UC region stimulated the formation of active IRF3 dimers and expression of ISG56, an IRF3 target gene (shown by immunoblotting)
PU/UC region forms a complex with RIG-I, while the X region does not (shown by a gelshift assay)
Induction of IFNβ promotor depends on
RIG-I
IFNβ promotor induction in
Huh7.5 cells
Huh7.5 cell lack functional
RIG-I
cells were refractory to HCV
RNA induced signalling, which was rescued by overexpression of WT RIG-I
Involvement of signalling molecules in IFNβ production
reporter assays in MEFs (mouse embryonic fibroblasts)
MDA5 is also a RLR member like RIG-I; MyD88 and TRIF are essential adaptor protein used by TLR 7/8 and 3 recognizing endosomal RNA
no reporter induction in RIGI negative MEF‘s upon RNA stimulation
lack of MDA5, MyD88 and Trif does not influence signal induction by full length 3‘ NTR and PU/UC region
HCV PU/UC region co-localizes and interacts with RIG-I
FRET analysis
PU/UC RNA co-localizes and interacts with RIG-I
RIG-I is the essential PRR that signal innate immune responses against HCV triggered by the poly (U/UC) region
HCV RNA requires 5‘ ppp for RIG-I binding and signal triggering
RIGI binds to PAMP RNA containing 5‘ terminal triphosphate (5‘ ppp)
5‘ ppp is required for poly (U/UC) RNA binding by RIG-I and for IFN-β signalling, but does not mediated binding of RIG-I to the X region; X region just weakly triggers signalling reporter assay in
Huh7 cells, with and without pre-treatement with IFNβ gel-shift assay
N = N-terminus of RIG-I
FL = full lenght RIG-I
Effect of PU/UC or X RNA on RIG-I activation
limited trypsin digestion analysis
upon binding of PAMP RNA the
RIG-I repressor domain (RD) is displaced and present as a single fragment
binding of PU/UC region to RIG-I rendered the RD fragment
HCV PU/UC region directs stable interaction with RIGI in a 5‘ ppp dependent manner to activate signalling
Effect of nucleotide composition on IFNβ promotor signalling
replacement of uridine reduced PAMP signalling
poly-A is also capable to induce signalling, again replacement leads to reduced signalling
truncation of the PU/UC region also reduces signalling (not shown)
Effect of nucleotide composition on RIG-I activation
poly-A RNA as well as PU/UC bind to RIG-I and lead to displacement of the
RD
signalling can be triggered by polymeric uridine and riboadenine motifs serving as PAMP signature within 5‘ ppp RNA recognized by RIG-I
Induction of IFNβ promotor by PU/UC in vivo
signalling analysis in WT and
RIG-I -/mice
intravenous administration of HCV
RNA
induction of hepatic IFNβ mRNA levels in WT mice, but not RIG-I -/mice, by HCV genome or PU/UC region
HCV PAMP RNA triggers hepatic immune responses (1)
time course studies
the PU/UC region induced a peak of hepatic IFNβ mRNA levels and
IFNβ serum levels in WT mice, but not in RIG-I-/- mice
HCV PAMP RNA triggers hepatic immune responses (2)
the PU/UC region induced also a peak of hepatic RIG-I and ISG56 mRNA levels in WT mice,but not in RIG-I-/- mice
induction of tissue-wide expression of ISG54 in WT mice suggestingthat paracrine signalling of IFNβ could play a role in hepatic defenses against
HCV
HCV PAMP RNA triggers paracrine antiviral effects of the innate immune response
measurement of HCV production of infected Huh7.5 cells treated with
IFNβ or conditioned media
(supernatant from cells transfected with the indicated RNA species)
treatment with IFNβ or supernatant from PU/UCtransfected cells induced a immune response that suppresses HCV infection
RIG-I signalling triggered by
PU/UC can induce an antiviral response through indirect, paracrine actions of IFN produced from HCV
PAMP signalling pre-treatment with
IFNβ or conditioned media treatment with IFNβ or conditioned media
48h post infection
Summary
RIG-I signals innate immune responses against HCV triggered by the poly
(U/UC) region of the 3‘ NTR of the HCV genome or ist replication intermediate
IFNβ production is induced in experiments in vitro and in vivo in response to HCV RNA
PAMP signature recognized by RIG-I is characterized by polymeric uridine and riboadenine motifs within 5‘ ppp RNA
5‘ppp on PAMP RNA is necessary but not sufficient for RIG-I binding
RIG-I signalling triggered by PU/UC can induce a direct as well as a indirect antiviral response through paracrine actions of IFN produced from HCV
PAMP signalling
Thanks for your attention !