Use of Elicitor Sets to Characterize Cellular Signal Transduction
advertisement

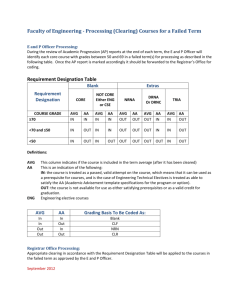
USE OF ELICITOR SETS TO CHARACTERIZE CELLULAR SIGNAL TRANSDUCTION Graduate Student: Arthi Narayanan Major Professor: Dr. Frank Chaplen Outline Background Experimental Methods Results & Discussion Background Complexities of signal transduction pathways What is systems biology? Does not investigate individual genes or proteins, but investigates the behavior and relationships of all of the elements in a particular biological system while it is functioning. Study of a biological system by a systematic and quantitative analysis of all of the components that constitute the system. Biological information has several important features: Operates on multiple hierarchical levels of organization. Processed in complex networks. Key nodes in the network where perturbations may have profound effects; these offer powerful targets for the understanding and manipulation of the system. Problem Statement Use the elicitor method - an experimental framework designed to monitor information flows through the G-protein signal transduction network. To derive mechanistic interpretations from the action of Phenylmethylsulfonyl Fluoride (PMSF), a serine protease inhibitor and nerve agent analog. Model System: Fish Chromatophores Overview of Chromatophores Aggregation/Dispersion of Fish Chromatophores Before and after 100 nM Clonidine Before and after 10 µM Forskolin Gq mediated signaling EXPERIMENTAL METHODS Elicitor sets method What is an elicitor panel? Known effectors of checkpoints in the signaling cascade. Elicitor = effector + application method Why elicitor sets? Enable identification of the key nodes in the signaling pathway Segregation of the pathway into different modules Perturbation of the signaling cascade by adding different effectors will help investigate the cross-talk mechanisms Enable signature identification of biologically active compounds A C B D 20-D mechanism space defined by elicitor panel described below and represented as 3-D projection (A) Cluster map for PMSF; (B) Cluster map for BC 1; (C) Cluster map for BC 5; (D) Cluster map for BC 6. The cluster map for each agent represents a unique complex signature defined by its biological mechanism of action. Elicitors are clonidine (100 and 50 nM), melanin stimulating hormone (10 nM) and forskolin (100 µM). Cross-talk between Gs and Gq pathways aq bg as bg AC PLC PLC IP3 cAMP RR Ca2+ PKA PKC DAG Cross-talk between Gi and Gq pathways bg aq ai bg AC PLC IP3 cAMP R Ca2+ PKA PKC DAG EXPERIMENTAL SET-UP Day 0: Plated cultured fish chromatophores in 24 well plates Day 1: Media change Day 2: Experiments Measured OD of cells at ground state Exposed cells to 10 µM forskolin for 24 minutes with OD being measured at regular intervals Added 1 mM PMSF to cells and measured OD values for 2.77 hours Added secondary elicitors (1&100 µM H89, 1&100 µM cirazoline, 100 nM clonidine) and monitored the response for 42 minutes. Plotted normalized % change in OD Vs Time RESULTS AND DISCUSSION Table 1: List of agents used with their concentrations and response patterns Concentration Point of action Response type Forskolin 10 µM Adenyl cyclase activator HyperDispersion PMSF 1 mM Serine protease Slight dispersion inhibitor at / d/s of PKA Clonodine 100 nM Gi activator Aggregation Cirazoline 1 & 100 µM Gq activator Aggregation H 89 1 & 100 µM PKA inhibitor Aggregation MSH 1 nM Gs activator Dispersion Optical density Dilution curves for Clonidine, Cirazoline and L-15 control 120 NORMALIZED % OD CHANGE 100 80 60 AVG:500 nm CLO AVG:100 nm CLO AVG:10 nm CLO AVG:1 um CRZ AVG:100 um CRZ AVG:10 um CRZ AVG:L-15 40 20 0 0 500 1000 1500 2000 TIME, SECONDS 2500 3000 3500 Dose response curves for H-89 and DMSO controls 120 NORMALIZED % OD CHANGE 100 80 AVG:10 nm H89 AVG:100 nm H89 60 AVG:1 um H89 AVG:10 um H89 AVG:100 um H89 40 AVG:DMSO control 100 nm AVG:DMSO control 10 nm AVG:DMSO control 1 um 20 AVG:DMSO control 10 um AVG:DMSO control 100 um 0 0 200 400 600 TIME, SECONDS 800 1000 1200 Dilution curves for Forskolin and MSH 160 NORMALIZED % OD CHANGE 140 120 AVG:100 um Fors 100 AVG:1 um Fors AVG:10 um Fors 80 AVG:10 nm MSH AVG:1 nm MSH 60 AVG:0.1 nm MSH 40 20 0 0 500 1000 1500 2000 TIME, SECONDS 2500 3000 3500 Segmentation of the cAMP pathway by application of forskolin as the primary elicitor 140 NORMALIZED % OD CHANGE 120 AVG: Fors_H89 1 uM 100 AVG: Fors_H89 100 uM AVG: Fors_CRZ 1 uM AVG: Fors_CRZ 100 uM 80 AVG: Fors_CLO 100 nM AVG: DMSO_H89 1 um 60 AVG: DMSO_H89 100 um AVG: DMSO_CRZ 1 um AVG: DMSO_CRZ 100 um 40 AVG: DMSO_CLO 100 nm 20 0 0 500 1000 1500 TIME, SECONDS 2000 2500 Experiments with MSH as the primary elicitor 140 NORMALIZED % OD CHANGE 120 100 AVG: MSH_H89 1uM AVG: MSH_H89 100 uM AVG: MSH_CRZ 1 uM 80 AVG: MSH_CRZ 100 uM AVG: MSH_CLO 100 nM AVG: L15_H89 1 uM 60 AVG: L15_H89 100 uM AVG: L15_CRZ 1 uM AVG: L15_CLO 100 nM 40 AVG: L15_CRZ 100 uM 20 0 0 500 1000 1500 TIME, SECONDS 2000 2500 Elicitor experiments with PMSF applied after forskolin 180 160 NORMALIZED % OD CHANGE 140 120 avg FOR_PMSF_1 um H89 100 avg FOR_PMSF_100 um H89 avg FOR_PMSF_1 um CRZ 80 avg FOR_PMSF_100 um CRZ avg FOR_PMSF_100 nm CLO 60 40 20 0 0 2000 4000 6000 8000 10000 TIME, SECONDS 12000 14000 16000 DMSO and Ethanol controls 220 200 NORMALIZED % OD CHANGE 180 160 avg FOR_EtOH_1 um CRZ avg FOR_EtOH_100 um CRZ 140 avg FOR_EtOH_100 nm CLO avg DMSO_EtOH_100 nm CLO 120 avg DMSO_EtOH_100 um CRZ avg DMSO_EtOH_1 um CRZ 100 avg DMSO_EtOH_1 um H89 80 avg DMSO_EtOH_100 um H89 avg FOR_EtOH_1 um H89 60 avg FOR_EtOH_100 um H89 40 20 0 0 2000 4000 6000 8000 10000 12000 14000 16000 TIME, SECONDS TARGETS FOR PRIMARY AND SECONDARY ELICITORS Clonidine Gq Gi Cirazoline PLC AC Forskolin PIP2 IP3 + DAG cAMP Ca++ H89 PKA PKC Aggregation Aggregation Mechanistic interpretation from PMSF action %OD change due to H-89 in: wells treated with PMSF - 26% control wells - 44% Our experimental results predict that PMSF acts at or downstream of PKA. An interpretation of the results suggests an interaction between a serine protease and PKA, that makes the latter less susceptible to H89. When PMSF, a serine protease inhibitor is added to the cells, this interaction is hampered thereby allowing H-89 to totally exert its inhibitory effect on PKA. Discussion and Conclusion Choice of AC as reference node and forskolin as primary elicitor simplifies the determination of the mechanism of action of PMSF. Application of PMSF after forskolin localized the measurable effect of PMSF to regions of the signaling cascade, below AC Perturbation by addition of secondary elicitors provided more information within the simplex scenario created by forskolin. Increased information resolution is evident in the heightened sensitivity of PKA to H-89 in the presence of PMSF, while the upper segment of the pathway is decoupled through application of forskolin help identify cross-talks. Failure of cirazoline to elicit a response when applied after forskolin shows an evidence of cross-talk. Thanks To: • Dr.Frank Chaplen for his indispensable support and guidance at every step during my research. • Dr. Rosalyn Upson for her guidance and encouragement. • Elena, Linda, June, Ruth, Christy, Bob and Indi for all your help along the way. • Dr.Michael Schimerlik and Dr. Skip Rochefort for serving on my committee. • Jeanine Lawrence, Ljiljana Mojovic and Ned Imming for your help in the lab. • Ganesh and my family back in India for everything. • NSF and AES for funding this work.