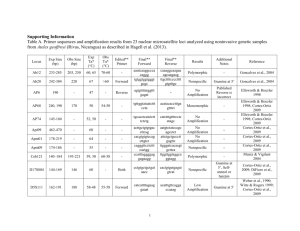
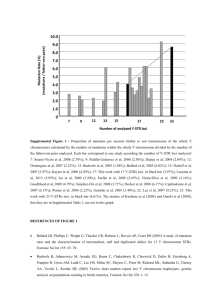
Présentation PowerPoint
advertisement

Melampsora Genome Consortium - 2008 Summer Workshop Microsatellites in the Mlp genome Pascal Frey, Constance Xhaard, Axelle Andrieux, Benoît Barrès, Fabien Halkett INRA Nancy Joint Research Unit « Tree – Microbe Interactions » Champenoux, France Reminder: what is a microsatellite? • Microsatellites = SSR (Simple Sequence Repeats) = VNTR (Variable Number of Tandem Repeats • Motifs : di-, tri-, tetra-, penta-, hexa-, or hepta-nucleotides e.g. (TA)n, (TCA)n, (CAGT)n • Highly variable genetic markers Very useful for population genetics, genetic mapping, diagnosis... Why studying microsatellites in the Mlp genome? 1. To determine the number and the distribution of microsatellite loci in the Mlp genome. 2. To build a genetic map of Mlp. Problems to obtain a large offspring population. 3. To develop new microsatellite loci for population genetic studies. Microsatellite finding with Magellan (1) Magellan software : Dee Carter, Univ. Sydney www.medfac.usyd.edu.au/people/academics/profiles/dcarter.php 1st run (march 2007) on the 1st assembly of the Mlp genome (Jazz) Genome size: 122 Mb; 2,682 scaffolds 7,530 microsatellite loci found (min 6 repeats for all motifs) 59% di, 34% tri, 3% tetra, 2% penta, 2% hexa-nucleotides Culling settings : min 10 repeats for di-nucl., and 8 repeats for tri-nucl. 664 microsatellite loci found 17% di, 39% tri, 17% tetra, 11% penta, 14% hexa-nucleotides Microsatellite finding with Magellan (2) 2nd run (July 2008) on the 2nd assembly of the Mlp genome (Arachne) Genome size: 102 Mb; 462 scaffolds 7,293 microsatellite loci found (min 6 repeats for all motifs) 58% di, 35% tri, 3% tetra, 2% penta, 2% hexa-nucleotides Culling settings : min 10 repeats for di-nucl., and 8 repeats for tri-nucl. 654 microsatellite loci found (652 in the main genome, 1 in the mitochondrial DNA, 1 in Alt-haplotypes, none in Repetitive elements) 18% di, 39% tri, 16% tetra, 12% penta, 13% hexa-nucleotides Microsatellite finding with Magellan (3) Comparison of both runs (and of both assemblies) Most of the microsatellite loci were found in both runs. In some cases, 2 microsatellite loci were defined on 2 separate scaffolds of the 1st assembly, and corresponded to only 1 locus in the 2nd assembly. -> both alleles for strain 98AG31 Validation of the microsatellites found (1) • Design of 1-2 primer pairs for each of the 650 loci with Primer3 software. • In vitro PCR assays to validate the loci: 75% amplification success • Determination of the polymorphism of the loci on 2 sexual populations of 40 Mlp isolates. • Suppression of some loci not at Hardy-Weinberg equilibrium. Validation of the microsatellites found (2) Microsatellite loci resolved on a Beckman CEQ8000 DNA sequencer homozygous loci heterozygous locus Validation of the microsatellites found (3) 40 35 30 Design of 2 multiplex panels 25 of 16 and 14 loci to be amplified in a single tube 20 15 10 5 0 50 150 250 350 450 550 650 Dye4 Dye3 -5 Dye2 NA -10 -15 Distribution of microsatellites on the 34 main scaffolds 4,5 4,0 Position (Mb) 3,5 3,0 2,5 2,0 1,5 1,0 0,5 0,0 0 5 10 15 20 25 30 Scaffold # Of the 462 scaffolds, only 111 contain microsatellites. Most of the smallest scaffolds contain no microsatellite. Average density: 6.5 microsatellites/Mb 35