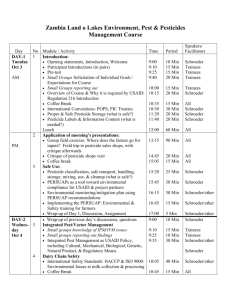
Database - BIOTEC - Biotechnology Center TU Dresden
advertisement
Introduction to Databases
Michael Schroeder
BioTechnological Center
TU Dresden
Biotec
Structure
Motivation
Introduction to MySQL
Example Queries
Using SQL to query SCOP
By Michael Schroeder, Biotec
2
Motivation
In the last term,
we accessed most information online via the web
we interacted directly and manually with databases and tools
we had to manually submit queries, interpret results. select interesting
results, cut&paste them, and submit queries again,…
Pro:
Reasonably easy to get hold of information
Con:
Not possible to ask many queries
Queries limited by interface provided by web page
Difficult/impossible to integrate information from different sites
In this term, we will look at the databases underlying the online
front ends
How is the data internally stored?
How can we - and more important computer programs - directly interact
with the underlying data, so that we can ask more powerful queries, large
queries, and integrate different systems
By Michael Schroeder, Biotec
3
What actually happens when you
retrieve data online?
LLNE
YLEEVE
EYEEDE
LLNE
YLEEVE
EYEEDE
Client
Message
Web Server
1
Get home page
2
Send home page
Get it and send it
3
Display home page, enter
query, and press submit
Send query
Start programme that
evaluates query by
accessing database…
4
Display result
Send result
… Compose result web
page and send it
By Michael Schroeder, Biotec
4
What actually happens
You are limited by what web server
allows you to ask:
Example CATH:
•PDB ID,
•CATH code, or
•General text
But you cannot ask:
•In how many different PDB
structures is there a P-loop domain?
•Is there a PDB entry with a P-loop
and a DNA-binding domain
•How many different superfamilies
does the largest structure in PDB
have?
•With direct access to the underlying
database you could answer all these
questions (and many more)
By Michael Schroeder, Biotec
5
Querying over the Web
Problem is always the same:
The web interface limits access to
the underlying database
How can we interact directly with
the database
By Michael Schroeder, Biotec
6
What databases are about
Logical organization of data
data models, schema design, dictionaries
Physical organization of data
Fast retrieval, indexing, compact storage of data
Other requirements:
Logging (important to know who did what to the data)
Security and access control (important to know who can
do what)
Transactions and concurrency control (important when
more than one person is working on database)
Integrity (important to ensure that only valid entries in the
database)
Recovery (important as hardware and software can
sometimes
fail
By Michael
Schroeder, Biotec
7
Different types of databases
Flat files
XML
Relational database
(Object databases)
(Object relational databases)
By Michael Schroeder, Biotec
8
Flat files
We can store any data in a flat
file, e.g. EMBL
But is this a database?
Logical data organisation:
None, unless we define one
(as done for EMBL) and adhere
to it, which is not enforced
Physical data organisation:
None, we cannot optimise
retrieval for common queries
Logging: No
Access control: Implicit
through Unix
Transaction and concurrency
control: None
Integrity: None
Recovery: If files are backedup they can be recovered.
However, not on the fly
By Michael Schroeder, Biotec
ID
XX
AC
XX
SV
XX
DT
DT
XX
DE
XX
KW
XX
OS
OC
OC
OC
XX
RN
RP
RX
RX
RA
RT
RT
RL
XX
RN
RX
RX
RA
RT
RT
BTBPTIG
standard; genomic DNA; MAM; 3998 BP.
X03365; K00966;
X03365.1
18-NOV-1986 (Rel. 10, Created)
20-MAY-1992 (Rel. 31, Last updated, Version 3)
Bovine pancreatic trypsin inhibitor (BPTI) gene
Alu-like repetitive sequence; protease inhibitor;
trypsin inhibitor.
Bos taurus (cow)
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata;
Euteleostomi; Mammalia;
Eutheria; Cetartiodactyla; Ruminantia; Pecora;
Bovoidea; Bovidae; Bovinae;
Bos.
[1]
1-3998
MEDLINE; 86158754.
PUBMED; 2420326.
Kingston I.B., Anderson S.;
"Sequences encoding two trypsin inhibitors occur in
strikingly similar
genomic environments";
Biochem. J. 233(2):443-450(1986).
[2]
MEDLINE; 84070725.
PUBMED; 6580617.
Anderson S., Kingston I.B.;
"Isolation of a genomic clone for bovine pancreatic
trypsin inhibitor by
using a unique-sequence synthetic DNA probe.";
9
XML files
We can store any data in XML,
the eXtentable Mark-up
Language, e.g. Medline
But is this a database?
Logical data organisation:
yes, XML schema, which is
enforced
Physical data organisation:
None, we cannot optimise
retrieval for common queries
Logging: No
Access control: Implicit
through Unix
Transaction and concurrency
control: None
Integrity: None
Recovery: If files are backedup they can be recovered.
However, not on the fly
By Michael Schroeder, Biotec
<Article>
<Journal>
<ISSN>0270-7306</ISSN>
<JournalIssue>
<Volume>19</Volume>
<Issue>11</Issue>
<PubDate>
<Year>1999</Year>
<Month>Nov</Month>
</PubDate>
</JournalIssue>
</Journal>
<ArticleTitle>Differential regulation of the cell wall
integrity mitogen-activated protein kinase pathway in
budding yeast by the protein tyrosine phosphatases
Ptp2 and Ptp3.
</ArticleTitle>
<Pagination>
<MedlinePgn>7651-60</MedlinePgn>
</Pagination>
<Abstract>
<AbstractText>Mitogen-activated protein kinases
(MAPKs) are inactivated by dual-specificity and
protein tyrosine phosphatases (PTPs) in yeasts. In
Saccharomyces cerevisiae, two PTPs, Ptp2 and
Ptp3, inactivate the MAPKs, Hog1 and Fus3, with
different specificities... </AbstractText>
</Abstract>
<Affiliation>Department of Chemistry, University of
Colorado, Boulder, Colorado 80309-0215, USA.
</Affiliation>…
10
Relational Database
Central Idea: Data as relations in a table
E.g. SCOP, Structural Classification of Proteins
+-------+------+---------+---------+--------------------------------------+
| id
| type | sccs
| sid
| description
|
+-------+------+---------+---------+--------------------------------------+
| 46457 | cf
| a.1
| | Globin-like
|
| 46458 | sf
| a.1.1
| | Globin-like
|
| 46459 | fa
| a.1.1.1 | | Truncated hemoglobin
|
| 46460 | dm
| a.1.1.1 | | Truncated hemoglobin
|
| 46461 | sp
| a.1.1.1 | | Ciliate (Paramecium caudatum)
|
| 14982 | px
| a.1.1.1 | d1dlwa_ | 1dlw A:
|
| 46462 | sp
| a.1.1.1 | | Green alga (Chlamydomonas eugametos) |
| 14983 | px
| a.1.1.1 | d1dlya_ | 1dly A:
|
| 63437 | sp
| a.1.1.1 | | Mycobacterium tuberculosis
|
| 62301 | px
| a.1.1.1 | d1idra_ | 1idr A:
|
+-------+------+---------+---------+--------------------------------------+
By Michael Schroeder, Biotec
11
Relational Database
Central Idea: Data as relations in a table
E.g. Employee
+-------+------+---------+---------+
| id
| name | salary | role
|
+-------+------+---------+---------+
| 46457 | pete | 50.000 | director|
| 46458 | jane | 60.000 | nurse
|
| 46459 | asif | 70.000 | driver |
+-------+------+---------+---------+
By Michael Schroeder, Biotec
12
Relational Database
Central Idea: Data as relations in a table
E.g. pets
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
13
Relational Database
Central Idea: Data as relations in a table
E.g. school
+-------+------+---------+
| id
| name | subject |
+-------+------+---------+
| 46458 | rick | bio
|
| 46459 | gerd | bio
|
| 46460 | mary | bio
|
| 46461 | ella | math
|
| 14982 | anne | math
|
| 46462 | paul | math
|
+-------+------+---------+
By Michael Schroeder, Biotec
+-------+------+---------+
| id
| prof | subject |
+-------+------+---------+
| 51221 | bert | bio
|
| 55435 | anne | math
|
+-------+------+---------+
+---------+------+-----+------+
| subject | room | day | time |
+---------+------+-----+------+
| bio
| A
| mo | 3pm |
| math
| B
| tue | 1pm |
+---------+------+-----+------+
14
Relational Database
A cell in the table stores a single number or string,
but not a list
+------+-------------+
| prof | subjects
|
+------+-------------+
| bert | {bio,sport} |
| anne | {math,arts} |
+------+-------------+
Lists, sets need to be flattened
+------+-------------+
| prof | subject
|
+------+-------------+
| bert | bio
|
| bert | sport
|
| anne | arts
|
| anne | math
|
+------+-------------+
By Michael Schroeder, Biotec
15
Bioinformatics: 10 years of
resistance to flattening!
Why the resistance?
Bioinformatics data is naturally nested
Extensive Use of sets and lists
E.g. Swissprot: Features, keywords, References
Such data can be flattened, but the resulting relational schema
is hard to understand hence it is hard to formulate queries.
For example, storing the SWISSPROT entry in a relational
database would split it over 15-20 tables.
By Michael Schroeder, Biotec
16
Relational Databases
RDB introduced in 1970 by Codd
Took off in the 80s
In the business world, relational databases are the rule
(Oracle, Sybase, mySQL, DB2, Microsoft Access).
Large biomedical databases typically use a relational
technology; but there are also a lot of homegrown
systems (ACeDB, SRS indexed files). Data is almost
always viewed and exported in a variety of flat file
formats (EMBL, GenBank among others)
By Michael Schroeder, Biotec
17
The flood of biomedical data…
Since 1980, the number and size of biomedical
databases has been growing exponentially.
How can you find sources of information you are
seeking?
Nucleic Acids Research Database Issue in January of
every year (http://nar.oupjournals.org/)
Dbcat (http://www.infobiogen.fr/ ): a flat file database
of 500 biological databases.
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
18
Relational Schema
The schema of a database is a set of relation names,
their field names and types.
Example:
Entry(ID: int, Length: int, Seq: string, Mod: date)
Feature(ID: int, Type: string, From: int,To: int)
Entry and Feature are relation names,
ID, Seq, Mod, etc are attribute names, and
int, string, date are domains
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
19
Relation Instance
An instance of a relation is a set of tuples of the type
of the relation.
A tuple of Entry could be:
( ID: 82814, Length: 597, Seq:“ccagctaaccg”, Mod: 1-7-95)
A tuple of Feature could be:
(ID: 82814, Type:“source”, From:1,To:8959)
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
20
Tabular representation
Typically, relations are displayed as tables
attributes
Sequence:
ID
82814
98608
Length
Seq
Mod
597
18976
“ccagctaa...”
“accgcct...”
1-07-95
2-14-98
tuples
Feature:
ID
Type
From
82814 “Source” 1
82814 “Gene”
23
By Michael Schroeder, Biotec
To
184
65
Susan B. Davidson, Biol537/CIS636, Fall 2003
21
Entities and Relationships
There is a one-many relationship from Entry to
Feature; each entry can have many features, but a
feature can be on at most one entry.
Put another way, the existence of a feature depends
on the existence of the owning entry referential
integrity
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
22
Integrity Constraints
ID is the key of Entry, indicated by underlining:
No two tuples of any instance of Entry can have the
same ID.
In Feature, there is a referential integrity constraint
on ID:
Every ID in Feature must appear in some tuple in
Entry.
This is specified in the data definition language
(DDL), and enforced by the system as updates are
made to the instance.
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
23
DDL for this relational schema
CREATE TABLE Entry
(Id INTEGER,
Length INTEGER,
Sequence LONGCHAR,
Mod DATE,
PRIMARY KEY (Id) )
By Michael Schroeder, Biotec
CREATE TABLE Feature
(Id INTEGER,
Type CHAR(15),
From INTEGER,
To INTEGER,
PRIMARY KEY (Id, Type,
From, To)
FOREIGN KEY (Id)
REFERENCES Entry
ON DELETE CASCADE
ON UPDATE CASCADE)
Susan B. Davidson, Biol537/CIS636, Fall 2003
24
Querying relational databases
The language SQL has become a standard for
querying relational databases. Based on a curious
mixture of the relational algebra and relational
calculus (formal languages), it allows new relations of
information to be computed from a set of relations.
Unlike the relational algebra, it allows other useful
stuff: count, sum, min, max, etc.
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
25
Basic Query
SELECT
FROM
WHERE
[DISTINCT] target-list
relation-list
qualification
relation-list A list of relation names (possibly with a
range-variable after each name).
target-list A list of attributes of relations in relation-list.
* can be used to denote all atts.
qualification Comparisons (Attr op const or Attr1 op
Attr2, where op is one of <, <=, >, >=, =, <>
combined using AND, OR and NOT.
DISTINCT (optional) keyword indicates that the
answer should not contain duplicates. Default is that
duplicates are not eliminated!
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
26
Conceptual Evaluation
Strategy
Compute the product of relation-list
Discard tuples that fail qualification
Project over attributes in target-list
If DISTINCT then eliminate duplicates
This is probably a very bad way of executing the query, and
a good query optimizer will use all sorts of tricks to find
efficient strategies to compute the same answer.
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
27
Sample tables
Sequence:
ID
Length
Seq
Mod
82814
98608
1
76582
597
18976
16665
9976
“ccagctaa...”
“accgcct...”
“gtgtaa….”
“actgga…”
1-07-95
2-14-98
1-19-97
2-29-00
Feature:
ID
82814
82814
1
13428
13428
By Michael Schroeder, Biotec
Type
“Source”
“Gene”
“Gene”
“Gene”
“Source”
From
1
23
3
11000
1
To
597
65
9999
16665
16665
Susan B. Davidson, Biol537/CIS636, Fall 2003
28
Simple queries
Print all sequences
SELECT *
with length less
FROM Sequence
WHERE Length < 10000; than 10000.
ID
82814
76582
Length
Seq
Mod
597
9976
“ccagctaa...”
“actgga…”
1-07-95
2-29-00
SELECT Type
FROM Feature;
Print the type of all
features.
By Michael Schroeder, Biotec
Type
“Source”
“Gene”
“Gene”
“Gene”
“Source”
Susan B. Davidson, Biol537/CIS636, Fall 2003
29
Distinct
Note that SQL did not eliminate duplicates. We
need to request this explicitly.
SELECT DISTINCT Type
FROM Feature;
Print the type of all
features
(no duplicates).
By Michael Schroeder, Biotec
Type
“Source”
“Gene”
Susan B. Davidson, Biol537/CIS636, Fall 2003
30
Pattern Matching
Can be used in where clause. “_” denotes any
character, “%” 0 or more characters.
SELECT *
FROM Sequence
WHERE Seq LIKE ‘a_%g'
ID
98608
76582
Length
Seq
Mod
18976
9976
“accgcct...”
“actgga…”
2-14-98
2-29-00
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
31
Arithmetic
“as” can be used to label columns in the output;
arithmetic can be used to compute results
SELECT DISTINCT ID, To-From+1 as Length
FROM Feature;
ID
82814
82814
13428
13428
13428
By Michael Schroeder, Biotec
Length
597
43
9997
5666
16665
Susan B. Davidson, Biol537/CIS636, Fall 2003
32
Set operations -- union
SELECT ID
FROM Sequence
WHERE Length<10000
UNION
SELECT ID
FROM Feature
WHERE Type=“Source”;
ID
76582
82814
13428
• Duplicates do not occur in the union.
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
33
The UNION ALL operator
preserves duplicates
SELECT ID
FROM Sequence
WHERE Length<10000
UNION ALL
SELECT ID
FROM Feature
WHERE Type=“Source”;
By Michael Schroeder, Biotec
ID
76582
82814
82814
13428
Susan B. Davidson, Biol537/CIS636, Fall 2003
34
Intersection and difference
SELECT Id
FROM Sequence
INTERSECT
SELECT Id
FROM Feature;
SELECT Id
FROM Sequence
MINUS
SELECT Id
FROM Feature;
ID
ID
82814
13428
98608
76582
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
35
Products
SELECT *
FROM Sequence,Feature;
ID
Length
82814
597
98608
18976
1
16665
76582
9976
……. (lots more!)
Seq
Mod
ID
“ccagctaa...”
“accgcct...”
“gtgtaa….”
“actgga…”
1-07-95
2-14-98
1-19-97
2-29-00
82814
82814
82814
82814
Type
From To
“Source”
“Source”
“Source”
“Source”
1
1
1
1
597
597
597
597
Note that the ID column name is duplicated in the output.
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
36
Conditional join
SELECT *
FROM Sequence, Feature
WHERE Sequence.Id = Feature.Id;
ID
Length
82814
82814
13428
1
13428
597
597
16665
16665
16665
Seq
Mod
“ccagctaa...”
“ccagctaa...”
“gtgtaa….”
“gtgtaa….”
“gtgtaa….”
By Michael Schroeder, Biotec
ID
1-07-95 82814
1-07-95 82814
1-19-97 13428
1-19-97 13428
1-19-97 13428
Susan B. Davidson, Biol537/CIS636, Fall 2003
Type
From To
“Source” 1
“Gene” 23
“Gene”
3
“Gene” 11000
“Source” 1
597
65
9999
16665
16665
37
Counting
Print the number
of feature entries.
SELECT COUNT(*)
FROM Feature;
Print the number
of types of features.
SELECT COUNT(Type)
FROM Feature;
Surprisingly, the answer to both of these is the
following:
COUNT(TYPE)
5
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
38
Counting, cont.
To fix this, we use the keyword “DISTINCT”:
SELECT COUNT(DISTINCT Type)
FROM Feature;
COUNT(DISTINCT Type)
3
Can also use SUM, AVG, MIN and MAX.
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
39
Group by
So far, these aggregate operators have been
applied to all qualifying tuples. Sometimes we
want to apply them to each of several groups of
tuples.
For example: “Print the type and number of
features of each type.”
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
40
Group by
SELECT Type, COUNT(*)
FROM Feature
GROUP BY Type;
Type COUNT(*)
“Source”
2
“Gene”
3
Note that only the columns that appear in the
GROUP BY statement and “aggregated” columns can
appear in the output. So the following would generate
an error.
SELECT Type, From, To, COUNT(*)
FROM Feature
GROUP BY Type;
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
41
Group by … having
HAVING is to GROUP BY as WHERE is to FROM
SELECT Type, COUNT(*)
FROM Feature
WHERE From-To > 50
GROUP BY Type
HAVING AVG(From-To)> 8500
TYPE
COUNT(*)
“Source”
2
“HAVING” is used to restrict the groups that appear in the
result.
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
42
Summary
SQL is “relationally complete”: allows you to perform
operators in an algebra of relations (the relational
algebra).
Additional features: string comparisons, set
membership, arithmetic and grouping.
In contrast, Entrez is a much more limited language.
By Michael Schroeder, Biotec
Susan B. Davidson, Biol537/CIS636, Fall 2003
43
A Little Exercise
By Michael Schroeder, Biotec
44
A Little Exercise
Given the table pet below let us formulate some
queries…
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
45
A Little Exercise
Get all pet names…
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
46
A Little Exercise
SELECT name
FROM pet;
Get all owners and list them only once
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
47
A Little Exercise
SELECT DISTINCT owner
FROM pet;
Select the names of all birds…
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
48
A Little Exercise
SELECT name
FROM pet
WHERE species=“bird”;
Select the names of all female birds…
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
49
A Little Exercise
SELECT name
FROM pet
WHERE species=“bird” AND sex=“f”;
Select names and owners
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
50
A Little Exercise
SELECT name,owner
FROM pet;
Select owners of birds and dogs…
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
51
A Little Exercise
SELECT owner
FROM pet
WHERE species=“bird” OR species=“dog”;
Select all owners starting with Dia…
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
52
A Little Exercise
SELECT owner
FROM pet
WHERE owner LIKE “Dia%”;
How many pets has Gwen?...
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
53
A Little Exercise
SELECT count(name)
FROM pet
WHERE owner=“Gwen”;
Select owners of male pets in sorted order…
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
54
A Little Exercise
SELECT owner
FROM pet
WHERE sex=“m”
ORDER BY owner;
List owners and the
number of pets they
have…
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
55
A Little Exercise
SELECT owner, COUNT(name)
FROM pet
GROUP BY owner;
List owners and
the number of
pets they have
in descending
order…
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
56
A Little Exercise
SELECT owner, COUNT(name) AS
num
FROM pet
GROUP BY owner
ORDER BY num DESC;
List owners and the
number of pets they
have in descending
order, but only if
they have more
than 1 pet…
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
57
A Little Exercise
SELECT owner, COUNT(name) AS num
FROM pet
GROUP BY owner HAVING num > 1
ORDER BY num DESC;
List all pairs of cats and dogs…
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
58
A Little Exercise
SELECT p1.name, p1.species, p2.name, p2.species
FROM pet AS p1, pet AS p2
WHERE p1.species=“dog” AND p2.species=“cat”;
Select all male/female pairs of the same species…
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
59
A Little Exercise
SELECT p1.name, p1.species, p1.sex, p2.name, p2.species,
p2.sex
FROM pet AS p1, pet AS p2
WHERE p1.species=p2.species AND p1.sex=“m” AND p2.sex=“f”;
Can we write p1.sex != p2.sex instead? …
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
60
A Little Exercise
We would get the pair Whistler and Chirpy as
well
+----------+--------+---------+------+------------+------------+
| name
| owner | species | sex | birth
| death
|
+----------+--------+---------+------+------------+------------+
| Whistler | Gwen
| bird
|
| 0000-00-00 | NULL
|
| Chirpy
| Gwen
| bird
| f
| 1998-09-11 | 0000-00-00 |
| Bowser
| Diane | dog
| m
| 1979-08-31 | 1995-07-29 |
| Fang
| Benny | dog
| m
| 1990-08-27 | 0000-00-00 |
| Buffy
| Harold | dog
| f
| 1989-05-13 | 0000-00-00 |
| Claws
| Gwen
| cat
| m
| 1994-03-17 | 0000-00-00 |
| Fluffy
| Harold | cat
| f
| 1993-02-04 | 0000-00-00 |
| Slim
| Benny | snake
| m
| 1996-04-29 | 0000-00-00 |
+----------+--------+---------+------+------------+------------+
By Michael Schroeder, Biotec
61
A Little Science
When working with SCOP through the web interface
we are limited in what we can ask
What can we get out of SCOP when it is available as
a relational table?
A reminder
Classes: all alpha, all beta, alpha/beta, alpha+beta
SCOP family: >30% sequence similarity
SCOP superfamily: good structural similiary (possibly
<30%)
By Michael Schroeder, Biotec
62
A Little Science
At low sequence identity, good structural
alignments possible
Family
Same
Superfamily,
But not family
30%
By Michael Schroeder, Biotec
Picture from www.jenner.ac.uk/YBF/DanielleTalbot.ppt
63
A Little Science
Three tables:
cla, PDB entry and reference to its class, fold,
superfamily, family, domain
des, description of each node in the SCOP hierarchy
subchain, chain and possibly beginning and end on
chain for a domain instance
astral, sequence for a domain
By Michael Schroeder, Biotec
64
A Little Science
mysql> SELECT * FROM cla LIMIT 1;
+---------+--------+---------+-------+-------+-------+-------+-------+-------+-------+
| sid
| pdb_id | sccs
| cl
| cf
| sf
| fa
| dm
| sp
| px
|
+---------+--------+---------+-------+-------+-------+-------+-------+-------+-------+
| d1dlwa_ | 1dlw
| a.1.1.1 | 46456 | 46457 | 46458 | 46459 | 46460 | 46461 | 14982 |
+---------+--------+---------+-------+-------+-------+-------+-------+-------+-------+
mysql> SELECT * FROM des LIMIT 1;
+-------+------+------+------+--------------------+
| id
| type | sccs | sid | description
|
+-------+------+------+------+--------------------+
| 46456 | cl
| a
| | All alpha proteins |
+-------+------+------+------+--------------------+
mysql> SELECT * FROM astral LIMIT 1;
+---------+---------+-----------------------------------------------------------+
| sid
| sccs
| seq
|
+---------+---------+-----------------------------------------------------------+
| d1dlwa_ | a.1.1.1 | slfeqlggqaavqavtaqfyaniqadatvatffngidmpnqtnktaaflcaalgg...|
+---------+---------+-----------------------------------------------------------+
mysql> SELECT * FROM subchain LIMIT 1;
+----+-------+----------+-------+------+
| id | px
| chain_id | begin | end |
+----+-------+----------+-------+------+
| 1 | 14982 | A
|
|
|
+----+-------+----------+-------+------+
By Michael Schroeder, Biotec
65
Entity relationship diagram for SCOP
Thanks to Boris VassilevT
A Little Science
How many nodes are there in the hierarchy of type
class, fold, superfamily, family?
des
+-------+------+------+------+--------------------+
| id
| type | sccs | sid | description
|
+-------+------+------+------+--------------------+
| 46456 | cl
| a
| | All alpha proteins |
+-------+------+------+------+--------------------+
By Michael Schroeder, Biotec
67
A Little Science
How many nodes are there in the hierarchy of type
class, fold, superfamily, family?
Let us first find out how these types are called:
SELECT DISTINCT type
FROM des;
Now let’s list them with the numbers
des
+-------+------+------+------+--------------------+
| id
| type | sccs | sid | description
|
+-------+------+------+------+--------------------+
| 46456 | cl
| a
| | All alpha proteins |
+-------+------+------+------+--------------------+
By Michael Schroeder, Biotec
68
A Little Science
How many nodes are there in the hierarchy of type
class, fold, superfamily, family?
SELECT type, COUNT(*) AS num
FROM des
GROUP BY type
ORDER BY num;
There are not that many more
families than superfamilies.
Which superfamily has the most
families
+------+-------+
| type | num
|
+------+-------+
| cl
|
11 |
| cf
|
854 |
| sf
| 1305 |
| fa
| 2156 |
| dm
| 4567 |
| sp
| 7111 |
| px
| 44327 |
+------+-------+
des
+-------+------+------+------+--------------------+
| id
| type | sccs | sid | description
|
+-------+------+------+------+--------------------+
| 46456 | cl
| a
| | All alpha proteins |
+-------+------+------+------+--------------------+
By Michael Schroeder, Biotec
69
A Little Science
Which superfamily has the most families?
SELECT des.sccs, des.description,
COUNT(DISTINCT cla.fa) AS num
FROM des, cla
WHERE des.id=cla.sf
GROUP BY cla.sf
ORDER BY num DESC;
+---------+---------------------------------------------------------+-----+
| sccs
| description
| num |
+---------+---------------------------------------------------------+-----+
| a.4.5
| "Winged helix" DNA-binding domain
| 35 |
| c.69.1 | alpha/beta-Hydrolases
| 23 |
| c.66.1 | S-adenosyl-L-methionine-dependent methyltransferases
| 20 |
| c.52.1 | Restriction endonuclease-like
| 19 |
| c.37.1 | P-loop containing nucleotide triphosphate hydrolases
| 18 |
| b.18.1 | Galactose-binding domain-like
| 15 |
| d.92.1 | Metalloproteases ("zincins"), catalytic domain
| 14 |
| b.29.1 | Concanavalin A-like lectins/glucanases
| 14 |
| f.2.1
| Membrane all-alpha
| 13 |
| c.47.1 | Thioredoxin-like
| 12 |
| c.68.1 | Nucleotide-diphospho-sugar transferases
| 12 |
| c.2.1
| NAD(P)-binding Rossmann-fold domains
| 11 |
| a.4.1
| Homeodomain-like
| 10 |
| b.40.4 | Nucleic acid-binding proteins
| 10 |
| a.118.1
| Schroeder,
ARM repeat
| 1070|
By Michael
Biotec
A Little Science
Which families does the DNA binding-domain superfamily
have?
mysql> SELECT * FROM cla LIMIT 1;
+---------+--------+---------+-------+-------+-------+-------+-------+-------+-------+
| sid
| pdb_id | sccs
| cl
| cf
| sf
| fa
| dm
| sp
| px
|
+---------+--------+---------+-------+-------+-------+-------+-------+-------+-------+
| d1dlwa_ | 1dlw
| a.1.1.1 | 46456 | 46457 | 46458 | 46459 | 46460 | 46461 | 14982 |
+---------+--------+---------+-------+-------+-------+-------+-------+-------+-------+
mysql> SELECT * FROM des LIMIT 1;
+-------+------+------+------+--------------------+
| id
| type | sccs | sid | description
|
+-------+------+------+------+--------------------+
| 46456 | cl
| a
| | All alpha proteins |
+-------+------+------+------+--------------------+
The sccs of the superfamily is a.4.5. Its families have
sccs a.4.5.1, a.4.5.2,…, so how can we list them?
By Michael Schroeder, Biotec
71
A Little Science
Which families does the DNA binding-domain superfamily
have?
SELECT DISTINCT sccs, description
FROM des
WHERE sccs LIKE “a.4.5%” AND type=“fa”
ORDER BY sccs;
| sccs
| description
+----------+-----------------------------------------------| a.4.5.1 | Biotin repressor-like
| a.4.5.10 | Replication initiation protein
| a.4.5.11 | Helicase DNA-binding domain
| a.4.5.12 | Restriction endonuclease FokI, N-terminal (recognition) domain
| a.4.5.13 | Histone H1/H5
| a.4.5.14 | Forkhead DNA-binding domain
| a.4.5.15 | DNA-binding domain from rap30
| a.4.5.16 | C-terminal domain of RPA32
| a.4.5.17 | Cell cycle transcription factor e2f-dp
| a.4.5.18 | The central core domain of TFIIE beta
| a.4.5.19 | Z-DNA binding domain
| a.4.5.2 | LexA repressor, N-terminal DNA-binding domain
| a.4.5.20 | P4 origin-binding domain-like
| a.4.5.21 | ets domain
| a.4.5.22 | Heat-shock transcription factor
| a.4.5.23 | Interferon regulatory factor
…
By Michael Schroeder, Biotec
72
A Little Science
Which families does the DNA binding-domain superfamily have?
...
| a.4.5.24
| a.4.5.25
| a.4.5.26
| a.4.5.27
| a.4.5.28
| a.4.5.29
| a.4.5.3
| a.4.5.30
| a.4.5.31
| a.4.5.32
| a.4.5.33
| a.4.5.34
| a.4.5.35
| a.4.5.4
| a.4.5.5
| a.4.5.6
| a.4.5.7
| a.4.5.8
| a.4.5.9
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Iron-dependent represor protein
Methionine aminopeptidase, insert domain
mu transposase, DNA-binding domain
TnsA endonuclease, C-terminal domain
MarR-like transcriptional regulators
Plant O-methyltransferase, N-terminal domain
Arginine repressor (ArgR), N-terminal DNA-binding domain
C-terminal domain of the rap74 subunit of TFIIF
DEP domain
Lrp/AsnC-like transcriptional regulator N-terminal domain
Thanscriptional regulator IclR, N-terminal domain
SCF ubiquitin ligase complex WHB domain
C-terminal fragment of elongation factor SelB
CAP C-terminal domain-like
ArsR-like transcriptional regulators
GntR-like transcriptional regulators
Replication terminator protein (RTP)
N-terminal domain of molybdate-dependent transcriptional regulator ModE
Transcription factor MotA, activation domain
By Michael Schroeder, Biotec
73
A Little Science
Which families does the DNA binding-domain superfamily
have?
Let’s find example pdb’s
mysql> SELECT DISTINCT pdb_id FROM cla WHERE sccs="a.4.5.1";
+--------+
| pdb_id |
+--------+
| 1bia
|
| 1hxd
|
| 1bib
|
| 1j5y
|
+--------+
mysql> SELECT DISTINCT pdb_id FROM cla WHERE sccs="a.4.5.2";
+--------+
| pdb_id |
+--------+
| 1jhf
|
| 1jhh
|
| 1lea
|
| 1leb
|
+--------+
By Michael Schroeder, Biotec
74
A Little Science
1jhf
By Michael Schroeder, Biotec
1bia
75
A Little Science
1cgp
a.4.5.4
By Michael Schroeder, Biotec
76
A Little Science
1b9n, a.4.5.8
1bm9, a.4.5.7
By Michael Schroeder, Biotec
Some more…
1f4k, which family?
1f4k, a.4.5.7
1smt, a.4.5.5
1hw1, a.4.5.6
77
A Little Science
How many percent of superfamilies have only 1
family, how many 2,… ?
By Michael Schroeder, Biotec
78
A Little Science
How many percent of superfamilies have only 1,2,3,… families?
First let’s deposit the result of the query that found the number of
families for each superfamily in a table
CREATE TABLE fa_freq AS
SELECT des.sccs, des.description,
COUNT(DISTINCT cla.fa) AS num
FROM des, cla
WHERE des.id=cla.sf
GROUP BY cla.sf
ORDER BY num desc;
Now we count how many superfamilies have 1,2,3,… families
SELECT num AS fa_per_sf, COUNT(*) AS freq
FROM fa_freq
GROUP BY num;
By Michael Schroeder, Biotec
79
A Little Science
many percent of superfamilies have only 1,2,3,… families?
+-----------+------+
SELECT COUNT(*)
| fa_per_sf | freq |
FROM fa_freq ;
+-----------+------+
+----------+
|
1 | 981 | | count(*) |
|
2 | 164 | +----------+
|
3 |
65 | |
1305 |
|
4 |
29 | +----------+
|
5 |
25 |
|
6 |
14 |
|
7 |
6 |
|
8 |
5 |
SELECT num AS fa_per_sf,
|
9 |
1 |
(COUNT(*)/1305) AS perc
|
10 |
3 |
FROM fa_freq
|
11 |
1 |
GROUP BY num;
|
12 |
2 |
|
13 |
1 |
|
14 |
2 |
|
15 |
1 |
|
18 |
1 |
|
19 |
1 |
|
20 |
1 |
|
23 |
1 |
|
35 |
1 |
+-----------+------+
By Michael Schroeder, Biotec
+-----------+------+
| fa_per_sf | perc |
+-----------+------+
|
1 | 0.75 |
|
2 | 0.13 |
|
3 | 0.05 |
|
4 | 0.02 |
|
5 | 0.02 |
|
6 | 0.01 |
|
7 | 0.00 |
|
8 | 0.00 |
|
9 | 0.00 |
|
10 | 0.00 |
|
11 | 0.00 |
|
12 | 0.00 |
|
13 | 0.00 |
|
14 | 0.00 |
|
15 | 0.00 |
|
18 | 0.00 |
|
19 | 0.00 |
|
20 | 0.00 |
|
23 | 0.00 |
|
35 | 0.00 |
+-----------+------+
80
A Little Science
How many percent of superfamilies have only 1,2,3,…
families?
This is interesting! For the majority of superfamilies
there is only one family!
What is the PDB structure with the largest number of
(distinct) superfamilies?
By Michael Schroeder, Biotec
81
A Little Science
at is the PDB structure with the largest number of (distinct) superfamilies?
SELECT pdb_id,
COUNT(sf) AS sf_num
FROM cla
GROUP BY pdb_id
ORDER BY sf_num DESC
LIMIT 10;
+--------+--------+
| pdb_id | sf_num |
+--------+--------+
| 1aon
|
49 |
| 1hto
|
48 |
| 1ir2
|
48 |
| 1htq
|
48 |
| 1der
|
42 |
| 1f49
|
40 |
| 1jyy
|
40 |
| 1gho
|
40 |
| 1jyz
|
40 |
| 1jz1
|
40 |
+--------+--------+
By Michael Schroeder, Biotec
SELECT pdb_id,
COUNT(DISTINCT sf)
AS distinct_sf_num
FROM cla
GROUP BY pdb_id
ORDER BY distinct_sf_num DESC
LIMIT 10;
+--------+-----------------+
| pdb_id | distinct_sf_num |
+--------+-----------------+
| 1m1k
|
23 |
| 1k9m
|
23 |
| 1kd1
|
23 |
| 1kqs
|
23 |
| 1jj2
|
23 |
| 1k8a
|
23 |
| 1ffk
|
22 |
| 1i96
|
21 |
| 1hnz
|
20 |
| 1hr0
|
20 |
+--------+-----------------+82
A structure
with
23 different
superfamilies
1k9m Co
Crystal
Structure Of
Tylosin
Bound To
The 50S
Ribosomal
Subunit Of
Haloarcula
Marismortui
Ribosome
By Michael Schroeder, Biotec
83
A Little Science
Now let’s plot how many PDBs have 1, 2, 3,… distinct
superfamilies
First of all let us put the result of the previous slide in a table
(note if the table already exists we have to erase it first: DROP
TABLE pdb_sf_num. But be careful using DROP
CREATE TABLE pdb_sf_num AS
SELECT pdb_id,
COUNT(DISTINCT sf) AS
distinct_sf_num
FROM cla
GROUP BY pdb_id
ORDER BY distinct_sf_num DESC;
Now let us count how many PDBs with 1,2,3,… distinct
superfamilies…
By Michael Schroeder, Biotec
84
A Little Science
How many PDBs have 1, 2, 3,… distinct superfamilies
SELECT distinct_sf_num, COUNT(pdb_id) AS num
FROM pdb_sf_num
GROUP BY distinct_sf_num
ORDER BY distinct_sf_num;
+-----------------+-------+
| distinct_sf_num | num
|
+-----------------+-------+
|
1 | 13960 |
|
2 | 2721 |
|
3 |
495 |
|
4 |
178 |
|
5 |
33 |
|
6 |
25 |
|
7 |
1 |
|
9 |
4 |
|
20 |
9 |
|
21 |
1 |
|
22 |
1 |
|
23 |
6 |
By Michael Schroeder, Biotec
+-----------------+-------+
85
A Little Science
Let’s do the same in percent
SELECT COUNT(DISTINCT pdb_id)
FROM cla;
+----------+
| count(*) |
+----------+
|
17434 |
+----------+
There are 17434 PDB IDs
By Michael Schroeder, Biotec
86
A Little Science
How many PDBs have 1, 2, 3,… distinct superfamilies
SELECT distinct_sf_num, COUNT(pdb_id)/17434 AS perc
FROM pdb_sf_num
GROUP BY distinct_sf_num
ORDER BY distinct_sf_num;
+-----------------+------+
| distinct_sf_num | perc |
+-----------------+------+
|
1 | 0.80 |
|
2 | 0.16 |
|
3 | 0.03 |
|
4 | 0.01 |
|
5 | 0.00 |
|
6 | 0.00 |
|
7 | 0.00 |
|
9 | 0.00 |
|
20 | 0.00 |
|
21 | 0.00 |
|
22 | 0.00 |
|
23 | 0.00 |
+-----------------+------+
80% of PDB entries consist only of one type of superfamily!
By Michael Schroeder, Biotec
87
A Little Science
What are the most popular superfamilies?
I.e. for which are there the most PDB entries
By Michael Schroeder, Biotec
88
A Little Science
What are the most popular superfamilies?
SELECT des.sccs, des.description,
COUNT(DISTINCT cla.pdb_id) AS num_of_pdb_ids
FROM cla,des
WHERE des.id=cla.sf
GROUP BY cla.sf
ORDER BY num_of_pdb_ids DESC
LIMIT 10;
+--------+------------------------------------------------------+----------------+
| sccs
| description
| num_of_pdb_ids |
+--------+------------------------------------------------------+----------------+
| b.1.1 | Immunoglobulin
|
823 |
| d.2.1 | Lysozyme-like
|
777 |
| b.47.1 | Trypsin-like serine proteases
|
649 |
| c.37.1 | P-loop containing nucleotide triphosphate hydrolases |
521 |
| c.2.1 | NAD(P)-binding Rossmann-fold domains
|
384 |
| a.1.1 | Globin-like
|
384 |
| c.1.8 | (Trans)glycosidases
|
332 |
| b.50.1 | Acid proteases
|
288 |
| b.29.1 | Concanavalin A-like lectins/glucanases
|
230 |
| c.47.1 | Thioredoxin-like
|
217 |
+--------+------------------------------------------------------+----------------+
By Michael Schroeder, Biotec
89
A Little Science
Are all superfamilies equally likely to co-occur?
Let us generate a co-occurrence map as an answer
Which superfamilies co-occur most frequently
Which superfamilies have the most co-occurrence partners
The co-occurrence map should consist of two tables
A table with PDB ID, superfamily 1, superfamily 2 (to avoid
repetition we will require that sf1 is alphabetically before sf2)
A table with superfamily 1 and 2 and the number of PDBs
containing this co-occurrence
By Michael Schroeder, Biotec
90
A Little Science
Co-occurrence map:
SELECT DISTINCT c1.pdb_id, c1.sf, c2.sf
FROM cla AS c1, cla AS c2
WHERE c1.pdb_id=c2.pdb_id AND c1.sf<c2.sf
LIMIT 10;
+--------+-------+-------+
| pdb_id | sf
| sf
|
+--------+-------+-------+
| 1cqx
| 46458 | 63380 |
| 1cqx
| 46458 | 52343 |
| 1gvh
| 46458 | 63380 |
| 1gvh
| 46458 | 52343 |
| 1b33
| 46458 | 54580 |
| 1qgw
| 46458 | 56568 |
| 1kf6
| 46548 | 46977 |
| 1kf6
| 46548 | 51905 |
| 1kf6
| 46548 | 54292 |
| 1kf6
| 46548 | 56425 |
+--------+-------+-------+
We are still missing the sf names, which we can get from the des
table
By Michael Schroeder, Biotec
91
A Little Science
Co-occurrence Map:
CREATE TABLE cooc AS
SELECT DISTINCT c1.pdb_id,
c1.sf AS sf1, d1.description AS sf1name,
c2.sf AS sf2, d2.description AS sf2name
FROM cla AS c1, cla AS c2, des AS d1, des AS d2
WHERE c1.pdb_id=c2.pdb_id AND c1.sf<c2.sf AND
c1.sf=d1.id AND c2.sf=d2.id;
+--------+-------+--------------------------+-------+--------------------------------------------------------------+
| pdb_id | sf1
| sf1name
| sf2
| sf2name
|
+--------+-------+--------------------------+-------+--------------------------------------------------------------+
| 1cqx
| 46458 | Globin-like
| 63380 | Riboflavin synthase domain-like
|
| 1cqx
| 46458 | Globin-like
| 52343 | Ferredoxin reductase-like, C-terminal NADP-linked domain
|
| 1gvh
| 46458 | Globin-like
| 63380 | Riboflavin synthase domain-like
|
| 1gvh
| 46458 | Globin-like
| 52343 | Ferredoxin reductase-like, C-terminal NADP-linked domain
|
| 1b33
| 46458 | Globin-like
| 54580 | Allophycocyanin linker chain (domain)
|
| 1qgw
| 46458 | Globin-like
| 56568 | Non-globular alpha+beta subunits of globular proteins
|
| 1kf6
| 46548 | alpha-helical ferredoxin | 46977 | Succinate dehydrogenase/fumarate reductase C-terminal domain |
| 1kf6
| 46548 | alpha-helical ferredoxin | 51905 | FAD/NAD(P)-binding domain
|
| 1kf6
| 46548 | alpha-helical ferredoxin | 54292 | 2Fe-2S ferredoxin-like
|
| 1kf6
| 46548 | alpha-helical ferredoxin | 56425 | Succinate dehydrogenase/fumarate reductase catalytic domain |
+--------+-------+--------------------------+-------+--------------------------------------------------------------+
Now let us count the distinct PDB IDs for each co-occurrence
By Michael Schroeder, Biotec
92
A Little Science
Number of instances in co-occurrence map
SELECT COUNT(DISTINCT pdb_id) AS num,
sf1, sf1name, sf2, sf2name
FROM cooc
GROUP BY sf1,sf2
ORDER BY num DESC
LIMIT 10;
+-----+-------+-----------------------------------------------+-------+-----------------------------------------------| num | sf1
| sf1name
| sf2
| sf2name
+-----+-------+-----------------------------------------------+-------+-----------------------------------------------| 137 | 48726 | Immunoglobulin
| 54452 | MHC antigen-recognition domain
| 125 | 51011 | alpha-Amylases, C-terminal beta-sheet domain | 51445 | (Trans)glycosidases
| 117 | 47616 | Glutathione S-transferases, C-terminal domain | 52833 | Thioredoxin-like
| 99 | 47802 | DNA polymerase beta, N-terminal domain-like
| 56699 | Nucleotidyltransferases
| 97 | 53098 | Ribonuclease H-like
| 56672 | DNA/RNApolymerases
| 74 | 51735 | NAD(P)-binding Rossmann-fold domains
| 55347 | Glyceraldehyde-3-phosphate dehydrogenase-like,
| 64 | 48726 | Immunoglobulin
| 51445 | (Trans)glycosidases
| 63 | 51905 | FAD/NAD(P)-binding domain
| 54373 | FAD-linked reductases, C-terminal domain
| 58 | 50203 | Bacterial enterotoxins
| 54334 | Superantigen toxins, C-terminal domain
| 55 | 48726 | Immunoglobulin
| 51011 | alpha-Amylases, C-terminal beta-sheet domain
+-----+-------+-----------------------------------------------+-------+------------------------------------------------
Is it valid to draw any conclusions from the above table of superfamily
co-occurrences with their frequencies?
We should be careful, as the number of co-occurrences may be biased
by the abundance of each superfamily,
e.g. immunoglobulin is very frequent in the PDB
By Michael Schroeder, Biotec
93
A Little Science
This is quite similar a problem to the generation of
substitution matrices (last term)
A little reminder…
By Michael Schroeder, Biotec
94
BLOSUM
A Little Science
BLOcks SUbstitution Matrix (based on BLOCKS database)
Generation of BLOSUM x
Group highly similar sequences and replace them by a
representative sequences.
Only consider sequences with no more than x % similarity
Align sequences (no gaps)
For any pair of amino acids a,b and for all columns c of the
alignment, let q(a,b) be the number of co-occurrences of a,b in
all columns c.
Let p(a) be the overall probability of a occurring
BLOSUM entry for a,b is log2 ( q(a,b) / ( p(a)*p(b) ) )
By Michael Schroeder, Biotec
95
A Little Science
“Normalised” number of instances in cooccurrence map
Ok, so to avoid bias we will compute the following
Logarithm of probability of sf1 and sf2 co-occurring /
(probability of sf1 * probability of sf2)
To compute
the co-occurrence probabilities we will count the frequency of
sf1 and sf2 co-occurring and divide this by the overall number
of co-occurrences and
the probability of a superfamily we will divide the frequency of
the superfamily by the overall number of superfamilies
To compute this let us put “freq of sf1 and sf2 co-occurring” into a
table cooc_pdb and “freq of sf” into a table sf_pdb
By Michael Schroeder, Biotec
96
A Little Science
Count PDBs for each co-occurrence
CREATE TABLE cooc_pdb AS
SELECT COUNT(DISTINCT pdb_id) AS num,
sf1, sf1name, sf2, sf2name
FROM cooc
GROUP BY sf1,sf2
ORDER BY num DESC;
Count PDBs for each superfamily
CREATE TABLE sf_pdb AS
SELECT des.id AS sf, des.description,
COUNT(DISTINCT cla.pdb_id) AS num_of_pdb_ids
FROM cla,des
WHERE des.id=cla.sf
GROUP BY cla.sf
ORDER BY num_of_pdb_ids DESC;
By Michael Schroeder, Biotec
97
A Little Science
Count overall number of PDBs for each co-occurrence
SELECT SUM(num) AS totalCoocFreq
FROM cooc_pdb;
+---------------+
| totalCoocFreq |
+---------------+
|
9813 |
+---------------+
Count PDBs for each superfamily
SELECT SUM(num_of_pdb_ids) AS total_num_of_pdb_ids
FROM sf_pdb;
+----------------------+
| total_num_of_pdb_ids |
+----------------------+
|
22318 |
+----------------------+
By Michael Schroeder, Biotec
98
A Little Science
Normalised co-occurrence map
CREATE TABLE normalised_cooc AS
SELECT LOG((cooc_pdb.num/9813)/
((sf_pdb1.num_of_pdb_ids/22318)*(sf_pdb2.num_of_pdb_ids/22318))) AS
val, cooc_pdb.sf1, cooc_pdb.sf1name, cooc_pdb.sf2, cooc_pdb.sf2name
FROM cooc_pdb, sf_pdb AS sf_pdb1, sf_pdb AS sf_pdb2
WHERE cooc_pdb.sf1=sf_pdb1.sf AND cooc_pdb.sf2=sf_pdb2.sf
ORDER BY val desc;
+-----------+----------------------------------------------+-------------------------------+
| val
| sf1name
| sf2name
|
+-----------+----------------------------------------------+-------------------------------+
| 10.834834 | Arp2/3 complex 21 kDa subunit ARPC3
| Arp2/3 complex 16 kDa subunit |
| 10.834834 | tRNA splicing endonuclease, C-terminal domain| tRNA splicing endonuclease Edn|
| 10.834834 | Cell-division inhibitor MinC, C-terminal doma| Cell-division inhibitor MinC, |
| 10.834834 | Tricorn protease N-terminal domain
| Tricorn protease N-terminal do|
| 10.834834 | L-fucose isomerase, C-terminal domain
| L-fucose isomerase, N-terminal|
| 10.834834 | Catalytic domain of malonyl-CoA ACP transacyl| Probable ACP-binding domain of|
| 10.834834 | Transcription factor IIA (TFIIA), N-terminal | Transcription factor IIA (TFII|
| 10.834834 | Glutamyl tRNA-reductase dimerization domain | Glutamyl tRNA-reductase cataly|
| 10.834834 | N-terminal domain of phosphatidylinositol tra| C-terminal domain of phosphati|
| 10.834834 | Rotavirus NSP2 fragment, C-terminal domain
| Rotavirus NSP2 fragment, N-ter|
| 10.834834 | Lipovitellin-phosvitin complex, superhelical | Lipovitellin-phosvitin complex|
| 10.834834 | Aminoimidazole ribonucleotide synthetase (Pur| Aminoimidazole ribonucleotide |
| 10.834834 | Colicin E3 translocation domain
| Colicin E3 receptor domain
|
| 10.834834 | Arp2/3 complex 21 kDa subunit ARPC3
| Arp2/3 complex subunits
|
| 10.834834 | Arp2/3 complex 16 kDa subunit ARPC5
| Arp2/3 complex subunits
|
| 10.834834 | Head domain of nucleotide exchange factor Grp| Coiled-coil domain of nucleoti|
| 10.834834 | YhbC-like, C-terminal domain
| YhbC-like, N-terminal domain |
| 10.141687 | TolB, C-terminal domain
| TolB, N-terminal domain
|
By Michael Schroeder, Biotec
99
+-----------+----------------------------------------------+-------------------------------+
A Little Science
Which superfamily has the most co-occurrence
partners?
By Michael Schroeder, Biotec
100
A Little Science
Which superfamily has the most co-occurrence partners?
CREATE TABLE cooc_partner AS
SELECT COUNT(DISTINCT c2.sf) AS distinctnum,
COUNT(c2.sf) AS num, des.sccs, des.description
FROM cla AS c1, cla AS c2, des
WHERE c1.pdb_id=c2.pdb_id AND des.id=c1.sf
GROUP BY c1.sf;
SELECT *
FROM cooc_partner
ORDER BY distinctnum DESC
LIMIT 10;
| distinctnum | num
| sccs
| description
|
+-------------+-------+--------+------------------+
|
74 | 19177 | b.1.1 | Immunoglobulin
|
68 | 5228 | c.37.1 | P-loop
|
67 | 1602 | b.40.4 | Nucleic acid-binding proteins
|
42 |
435 | c.55.4 | Translational machinery components
|
42 |
580 | g.39.1 | Glucocorticoid receptor-like
|
31 |
561 | b.43.3 | Translation proteins
|
28 | 1575 | b.47.1 | Trypsin-like serine proteases
|
28 |
725 | d.14.1 | Ribosomal protein S5 domain 2-like
|
25 |
982 | a.4.5 | "Winged helix" DNA-binding domain
|
25 |
283 | d.52.3 | Prokaryotic type KH domain (pKH-…
+-------------+-------+--------+------------------+
By Michael Schroeder, Biotec
101
A Little Science
Which superfamily has the most co-occurrence partners?
SELECT *
FROM cooc_partner
ORDER BY num DESC
LIMIT 10;
+-------------+-------+---------+--------------+
| distinctnum | num
| sccs
| description
+-------------+-------+---------+--------------+
|
74 | 19177 | b.1.1
| Immunoglobulin
|
4 | 7532 | b.1.4
| beta-Galactosidase/glucuronidase
|
17 | 6355 | c.2.1
| NAD(P)-binding Rossmann-fold domains
|
24 | 6233 | f.2.1
| Membrane all-alpha
|
7 | 6007 | d.153.1 | N-terminal nucleophile aminohydrolases
|
1 | 5687 | i.1.1
| Ribosome and ribosomal fragments
|
68 | 5228 | c.37.1 | P-loop
|
18 | 5154 | c.1.8
| (Trans)glycosidases
|
5 | 3915 | a.1.1
| Globin-like
|
11 | 3880 | b.18.1 | Galactose-binding domain-like
+-------------+-------+---------+---------------+
By Michael Schroeder, Biotec
102
Scale-free Networks
Small-world property
“everybody on earth is related with a degree of six
intermediaries”
Reason: Network structure
A few highly connected nodes
Many nodes with few connections
Consequence: very short average distance between any
two nodes
Formally: Number of interaction partners follows powerlaw, i.e. distribution of number of interaction partners is
an exponential function
Protein interactions are small-world networks!
By Michael Schroeder, Biotec
103
Scale-free Networks
Is our co-occurrence map a scale-free network?
By Michael Schroeder, Biotec
104
Scale-free Networks
Is our co-occurrence map a scalefree network?
Let’s get the distribution of the
number of distinct co-occurrence
partners
SELECT distinctnum, COUNT(*)
FROM cooc_partner
GROUP BY distinctnum
ORDER BY distinctnum;
Let’s plot it
By Michael Schroeder, Biotec
+-------------+----------+
| distinctnum | count(*) |
+-------------+----------+
|
1 |
586 |
|
2 |
291 |
|
3 |
152 |
|
4 |
72 |
|
5 |
47 |
|
6 |
27 |
|
7 |
26 |
|
8 |
9 |
|
9 |
16 |
|
10 |
8 |
|
11 |
9 |
|
12 |
3 |
|
13 |
3 |
|
14 |
1 |
|
15 |
1 |
|
16 |
2 |
|
17 |
1 |
|
18 |
3 |
|
20 |
1 |
|
21 |
16 |
|
23 |
18 |
|
24 |
1 |
|
25 |
4 |
|
28 |
2 |
|
31 |
1 |
|
42 |
2 |
|
67 |
1 |
|
68 |
1 |
105
|
74 |
1 |
Scale-free Networks
Distribution of number of co-occurrence partners
(x-axis = number of partners, y-axis=frequency
By Michael Schroeder, Biotec
106
Scale-free Networks
But maybe the number of co-occurrence partners
is simply correlated to the number of PDB entries
we have for that superfamily, in which case the
power-law relationship would be an artefact
Let’s test this by extending the previous table with a
column for the average number of PDBs given as the
num column in cooc_partner
By Michael Schroeder, Biotec
107
Scale-free
Networks
Distinct number of cooccurrence partners and
average number of cooccurrences
SELECT distinctnum,
COUNT(*),AVG(num)
FROM cooc_partner
GROUP BY distinctnum
ORDER BY distinctnum;
By Michael Schroeder, Biotec
+-------------+----------+------------+
| distinctnum | count(*) | avg(num)
|
+-------------+----------+------------+
|
1 |
586 |
66.8328 |
|
2 |
291 |
124.0481 |
|
3 |
152 |
251.1316 |
|
4 |
72 |
324.1389 |
|
5 |
47 |
344.2340 |
|
6 |
27 |
289.7778 |
|
7 |
26 |
883.1923 |
|
8 |
9 |
663.8889 |
|
9 |
16 |
375.0625 |
|
10 |
8 |
772.3750 |
|
11 |
9 | 1454.3333 |
|
12 |
3 |
726.6667 |
|
13 |
3 |
642.3333 |
|
14 |
1 |
489.0000 |
|
15 |
1 | 2530.0000 |
|
16 |
2 |
645.0000 |
|
17 |
1 | 6355.0000 |
|
18 |
3 | 2254.3333 |
|
20 |
1 |
979.0000 |
|
21 |
16 |
254.1875 |
|
23 |
18 |
290.2778 |
|
24 |
1 | 6233.0000 |
|
25 |
4 |
433.0000 |
|
28 |
2 | 1150.0000 |
|
31 |
1 |
561.0000 |
|
42 |
2 |
507.5000 |
|
67 |
1 | 1602.0000 |
|
68 |
1 | 5228.0000 |
|
74 |
1 | 19177.0000
|
108
Scale-free Networks
No good correlation between frequency of number of distinct
co-occurrence partners and number of underlying instances.
Hence: scale-free property appears to be no artefact
By Michael Schroeder, Biotec
109
Limits
List all superfamily pairs which can interact directly or
indirectly (“Transitive Closure”).
This query cannot be expressed in SQL
SQL does not have the same “power” as a
programming language like Python
By Michael Schroeder, Biotec
110
Introduction to MySQL
By Michael Schroeder, Biotec
111
SQL: “Structured Query Language”—the most common
standardized language used to access databases.
SQL has several parts:
DDL – Data Definition Language
{Defining, Deleting, Modifying relation schemas}
DML – Data Manipulation Language
{Inserting, Deleting, Modifying tuples in database}
Embedded SQL – defines how SQL statements can be used
with general-purposed programming
By Michael Schroeder, Biotec
112
MySQL, the most popular Open Source SQL database, is
developed, distributed and supported by MySQL AB.
MySQL is a relational database management system.
MySQL software is Open Source.
• Written in C and C++. Tested with a broad range of different
compilers.
Works on many different platforms.
APIs for C, C++, Eiffel, Java, Perl, PHP, Python, Ruby,
and Tcl.
• You can find MySQl manual and documentation at:
www .mysql.com/documentation/
• You can download and install MySQL on your own computer
(both under Windows and Linux)
By Michael Schroeder, Biotec
113
MySQL
To see a list of options provided by mysql, invoke it with the --help option:
shell> mysql --help
Using SQL:
On any linux you have to use this to log on to MySQL:
shell> /usr/local/mysql/bin/mysql -h hostname -D loginname -p
shell> mysql -h host -u user -p
Enter password: ********
The ******** represents your password; enter it when
mysql displays the Enter password: prompt.
By Michael Schroeder, Biotec
114
Basic Query:
select A1, A2,…,An
from r1, r2, …,rm
where P;
A1, A2,…,An represent attributes
r1, r2, …rm represent relations
P represents predicate (guard condition)
Keywords may be entered in any letter case:
mysql> SELECT VERSION(), CURRENT_DATE;
mysql> select version(), current_date;
mysql> SeLeCt vErSiOn(), current_DATE;
By Michael Schroeder, Biotec
115
Prompt
Meaning
mysql>
->
‘>
Ready for new command.
Waiting for next line of multiple-line command.
Waiting for next line, collecting a string that
begins with a single quote (` ’ ’).
“>
Waiting for next line, collecting a string that
begins with a double quote (` ” ’).
mysql>
->
->
SELECT *
FROM my_table
WHERE name = “Smith” AND age < 30;
mysql> SELECT * FROM my_table WHERE name = "Smith AND
age < 30;
"> "\c
mysql>
\c to cancel the execution of a command
By Michael Schroeder, Biotec
116
Basic Database Operation
Create a database
Create a table
Load data into the table
Retrieve data from the table in various ways
Use multiple tables
Suppose you have several pets in your home (your menagerie) and you'd
like to keep track of various types of information about them. You can do so
by creating tables to hold your data and loading them with the desired
information. Then you can answer different sorts of questions about your
animals by retrieving data from the tables.
By Michael Schroeder, Biotec
117
Creating and Using a Database
mysql> SHOW DATABASES;
SHOW statement can be used to find out the databases currently
existing on the server
mysql> USE testdb
testdb is a database name.
USE command does not need a semi colon and must be given in a
single line.
Database needs to be invoked in order to use it.
mysql> CREATE DATABASE example;
Database names are case-sensitive unlike keywords; Same applies for
table names
So example != Example != EXAMPLE or some other variant
By Michael Schroeder, Biotec
118
Creating a Table
mysql> SHOW TABLES;
Displays the current list of tables
mysql> CREATE TABLE pet (name VARCHAR(20), owner VARCHAR(20),
-> species VARCHAR(20), sex CHAR(1), birth DATE, death DATE);
mysql> SHOW TABLES;
Will display the table with the table name pet
Verification of the table can be done with DESCRIBE command
mysql> DESCRIBE pet;
+---------+-------------+---------------+---------+---------+---------+
| Field
| Type
| Null
| Key
| Default | Extra
|
+---------+-------------+---------------+---------+---------+---------+
| name
| varchar(20) | YES |
| NULL
|
|
| owner
| varchar(20) | YES |
| NULL
|
|
| species | varchar(20) | YES |
| NULL
|
|
| sex
| char(1)
| YES |
| NULL
|
|
| birth
| date
| YES |
| NULL
|
|
| death
| date
| YES |
| NULL
|
|
By Michael Schroeder, Biotec
119
Loading Data into a Table
LOAD DATA uses a text file with single record in a line that match the attributes in the table.
Useful for inserting when multiple records are involved.
Example: pet.txt is a text file with a single record
Name
Whistler
owner
Gwen
species
bird
sex
\N
birth
1997-12-09
death
\N
mysql> LOAD DATA LOCAL INFILE "pet.txt" INTO TABLE pet;
INSERT command can be used when records needs to be inserted one at a time.
NULL can be directly inserted in the field column
Example:
mysql> INSERT INTO pet
-> VALUES ('Puffball','Diane','hamster','f','1999-03-30',NULL);
By Michael Schroeder, Biotec
120
Retrieving Information from a Table
The SELECT statement is used to pull information from a table. The general form of the
statement is:
SELECT what_to_select
FROM which_table
WHERE conditions_to_satisfy
The simplest form of SELECT retrieves everything from a table:
mysql> SELECT * FROM pet;
You can select only particular rows from your table.
mysql> SELECT * FROM pet WHERE name = "Bowser";
You can specify conditions on any column, not just name. For example, if you want to
know which animals were born after 1998, test the birth column:
mysql> SELECT * FROM pet WHERE birth >= "1998-1-1";
You can combine conditions, for example, to locate female dogs:
mysql> SELECT * FROM pet WHERE species = "dog" AND sex = "f";
By Michael Schroeder, Biotec
121
Selecting Particular Columns
If you don't want to see entire rows from your table, just name the columns in
which you're interested, separated by commas.
For example, if you want to know when your animals were born, select
the name and birth columns:
mysql> SELECT name, birth FROM pet;
To find out who owns pets, use this query:
mysql> SELECT owner FROM pet;
mysql> SELECT DISTINCT owner FROM pet;
You can use a WHERE clause to combine row selection with column selection.
For example, to get birth dates for dogs and cats only, use this query:
mysql> SELECT name, species, birth FROM pet
-> WHERE species = "dog" OR species = "cat";
By Michael Schroeder, Biotec
122
Sorting Rows
To sort a result, use an ORDER BY clause.
Here are animal birthdays, sorted by date:
mysql> SELECT name, birth FROM pet ORDER BY birth;
To sort in reverse order, add the DESC (descending) keyword to the name of
the column you are sorting by:
mysql> SELECT name, birth FROM pet ORDER BY birth DESC;
You can sort on multiple columns. For example, to sort by type of animal, then
by birth date within animal type with youngest animals first, use the
following query:
mysql> SELECT name, species, birth FROM pet ORDER BY species, birth
DESC;
By Michael Schroeder, Biotec
123
Pattern Matching
MySQL provides standard SQL pattern matching as well as a form of pattern
matching based on extended regular expressions similar to those used by Unix
utilities such as grep.
SQL pattern matching allows you to use `_' to match any single character and `%' to
match an arbitrary number of characters (including zero characters). In MySQL, SQL
patterns are case-insensitive by default. Some examples are shown here. Note that
you do not use = or <> when you use SQL patterns; use the LIKE or NOT LIKE
comparison operators instead.
To find names beginning with `b':
mysql> SELECT * FROM pet WHERE name LIKE "b%";
To find names containing exactly five characters, use the `_' pattern character:
mysql> SELECT * FROM pet WHERE name LIKE “_____”;
By Michael Schroeder, Biotec
124
Counting Rows
For example, you might want to know how many pets each owner has,
Counting the total number of animals you have is the same question as “How many rows
are in the pet table?”
The COUNT() function counts the number of non-NULL results, so the query to count your
animals looks like this:
mysql> SELECT COUNT(*) FROM pet;
You can use COUNT() if you want to find out how many pets each owner has:
mysql> SELECT owner, COUNT(*) FROM pet GROUP BY owner;
+---------+---------------+
| owner
| COUNT(*)
|
+---------+---------------+
| Benny
|
2
|
| Diane
|
2
|
| Gwen
|
3
|
| Harold |
2
|
+---------+---------------+
By Michael Schroeder, Biotec
125
Examples of some common queries
CREATE TABLE shop (
article
INT(4) UNSIGNED ZEROFILL DEFAULT
dealer
CHAR(20)
DEFAULT
price
DOUBLE(16,2)
DEFAULT
PRIMARY KEY(article, dealer));
‘0000'
‘’
'0.00’
NOT NULL,
NOT NULL,
NOT NULL,
INSERT INTO shop VALUES (1,'A',3.45),(1,'B',3.99),(2,'A',10.99),(3,'B',1.45),(3,'C',1.69),
(3,'D',1.25),(4,'D',19.95);
mysql> SELECT * FROM shop;
+--------------+---------+--------+
| article
| dealer | price |
+--------------+---------+--------+
|
0001 |
A | 3.45 |
|
0001 |
B | 3.99 |
|
0002 |
A | 10.99 |
|
0003 |
B | 1.45 |
|
0003 |
C | 1.69 |
|
0003 |
D | 1.25 |
|
0004 |
D | 19.95 |
+--------------+---------+--------+
The maximum value for a column
The row holding the maximum of a certain column
Maximum of column per group
The rows
holding
the group-wise
maximum of a certain field
By Michael
Schroeder,
Biotec
126
“What's the highest price?”
SELECT MAX(price) AS price FROM shop;
+---------+
| price |
+---------+
| 19.95 |
+---------+
“Find number, dealer, and price of the most expensive article.”
In ANSI SQL (and MySQL Version 4.1) this is easily done with a subquery:
SELECT article, dealer, price FROM shop
WHERE price = (SELECT MAX(price) FROM shop)
In MySQL versions prior to 4.1, you have to do it in two steps:
1. Get the maximum price value from the table with a SELECT statement.
2. Using this value compile the actual query:
SELECT article, dealer, price FROM shop WHERE price=19.95
Or, Using User Variables ( @variable-name, @temp := 5 )
Select @max_price := max(price) from shop;
Select article, dealer, price from shop where price = @max_price;
By Michael Schroeder, Biotec
127
Maximum of Column per Group
“What's the highest price per article?”
SELECT article, MAX(price) AS price
FROM shop
GROUP BY article;
+---------+---------+
| article | price |
+---------+---------+
| 0001 | 3.99 |
| 0002 | 10.99 |
| 0003 | 1.69 |
| 0004 | 19.95 |
+---------+---------+
By Michael Schroeder, Biotec
128
The Rows Holding the Group-wise Maximum of a Certain Field
“For each article, find the dealer(s) with the most expensive price.”
In ANSI SQL (MySQL Version 4.1 or greater), do it with a subquery
SELECT article, dealer, price
FROM shop s1
WHERE price=(SELECT MAX(s2.price)
FROM shop s2
WHERE s1.article = s2.article);
By Michael Schroeder, Biotec
129
But, In MySQL versions prior to 4.1, it has to be done in several steps, with a
temporary table (It doesn’t support nested-query \subquery).
CREATE TEMPORARY TABLE tmp
( article INT(4) UNSIGNED ZEROFILL DEFAULT '0000' NOT NULL,
price DOUBLE(16,2) DEFAULT '0.00' NOT NULL);
LOCK TABLES shop read;
INSERT INTO tmp
SELECT article, MAX(price) FROM shop GROUP BY article;
SELECT shop.article, dealer, shop.price
FROM shop, tmp
WHERE shop.article=tmp.article AND shop.price=tmp.price;
UNLOCK TABLES; DROP TABLE tmp;
By Michael Schroeder, Biotec
130