Presentation
advertisement

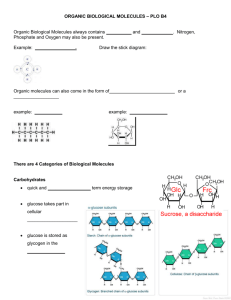
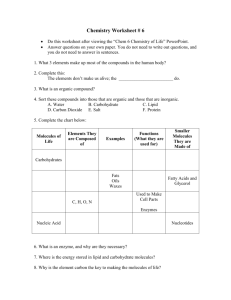
The Silicon Chemist: Using logic-based machine learning and virtual chemistry to design new drugs automatically. Christopher Reynolds and Mike Sternberg Predicted pKa Observed activity New drug leads are always needed, and virtual screening is now often used to simulate the bioactivity of compounds, in order to search as much of chemical space as possible in a reasonable amount of time. This project takes an existing logic based machine-learning approach to identifying bioactive compounds, and incorporates chemical synthesis rules, to design novel, easily synthesisable, and effective pharmaceutical drugs. True positives True negatives False negatives Fragmentation of molecules Figure 3. A scatter plot showing observed vs. predicted pKa in a cross validation of a PubChem bioassay. This shows that there are no false positives when using an activity cut-off of pKa 7. Top 1% (6 actives) Top 5% (25 actives) Top 10% (47 actives) Active/Inactive (232 actives) Observed pKa active(A):- positive(A, B), Nsp2(A, C), distance(A, B, C, 2.49, 0.5). Logic-based drug discovery • Inductive Logic Programming (ILP) is a machine learning technique, which learns human interpretable qualitative rules from chemical knowledge of active drugs (see Figure 2), which relate structure to activity, and can guide the next steps drug design chemistry. • Using Partial Least-Squares or Support Vector Machines, the rules can then be weighted • Weighted rules used as a quantitative model of Quantitative Structure-Activity Relationship (QSAR) to predict drug activity. • This approach combines the human comprehensible rules of ILP with the predictive accuracy of advanced regression techniques. • This Support Vector Inductive Logic Programming (SVILP) method is being patented. • INDDEx (Investigational Novel Drug Discovery by Example) is a proprietary virtual screening program, incorporating SVILP, and developed by Equinox Pharma Ltd., an Imperial College spinoff company. • The QSAR model is then used to screen a database of all purchasable molecules to identify drug leads. • In a blind test, INDDEx had a hit rate of 30%, predicting around 30 active molecules, each capable of being the start of a new drug series, and each sufficiently novel that it could be patented. False positives Department of Computing, Imperial College, and Department of Life Sciences, Imperial College True positive rate Introduction Results Database of virtual reactions Molecule is active if there is positive charge centre and an sp2 nitrogen atom 2.49±0.5Å. apart SVILP generates QSAR rules active(A):- phenyl(A, B), phenyl(A, C), distance(A, B, C, 4.1, 0.5). Molecule is active if there are two phenyl rings 4.1±0.5Å apart. Figure 2. Example ILP QSAR rules. Modified molecules False positive rate Figure 4. Receiver Operating Characteristic (ROC) curve, showing fraction of true positives against true negatives retrieved as the discrimination threshold is varied. ROC values for 1%, 5%, 10% and all actives are 0.912, 0.830, 0.814, and 0.892 respectively. This illustrates the sensitivity of active compound detection to hold true up to the top 5% most active compounds provided to INDDEx as examples. Screen Molecular database Screen Split molecule with reverse reaction Hit rate 30% Novel verified hits Modify using all viable reactions Novel verified hits on synthesisable molecules Figure 5. From left to right: wireframe visualisations of the two Hit rate reactant molecules entered in ?% SMILES format are processed by the SMIRKS esterification reaction, and the resultant product molecule is returned. Virtual chemistry To extend the machine learning method currently used in INDDEx, and move the process from hit to lead discovery, the Figure 1. Graphic showing the current method of logic-based rules produced by SVILP will be used to modify finding matches for ILP derived rules, as used by promising hits. This new process is shown in Figure 1. This the INDDEx software, and this project’s new project will concentrate on scanning through the dataset of method. purchasable molecules that partially fulfil the rules, and then altering the molecules to try and fit the remaining rules using a database of virtual chemical synthesis reactions to combine these molecules with a database of fragment-like molecules. Through this method, the program will generate focussed libraries of synthetic derivatives around the promising hit molecules, and so explore a far greater section of easily synthetically accessible chemical space.