Figure 4-1. Electron micrograph of a prokaryote, Bacillus subtilis
advertisement

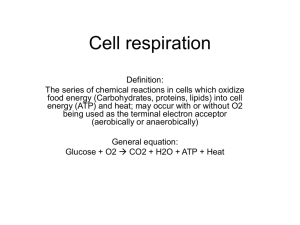
Environmental Biology for Engineers and Scientists D.A. Vaccari, P.F. Strom, and J.E. Alleman © John Wiley & Sons, 2005 Chapter 4 – The Cell Chapter 5 – Energy and Metabolism Figure 4-1. Example prokaryotic cell structure cell membrane ribosomes cell wall cytoplasm flagellum granules nucleoid thylakoid membrane Figure 4-2. Diagram of a plant cell and an animal cell. [From Schaum’s Biology] Figure 4-3. Plasma membrane structure. [Schaum’s “Human Anatomy and Physiology”] Figure 4-4. Chromosome structure in eukaryotes. [Based on P&H.] Scaffolding protein Chromosome Helical coil Histone protein DNA double helix Figure 4-5. The stages of mitotic division [Schaum’s Biology] Figure 4-6. Example life cycle of sexual organisms. Mitosis SOMATIC CELLS Diploid GERM CELLS Diploid Mitosis Meiosis ZYGOTE Diploid GAMETES Haploid Fertilization Figure 4-7. Meiosis: Crossing over and independent assortment of chromosomes. Interphase: replication One diploid cell Meiosis I: crossing over and first division Meiosis II: alignment and second division Four haploid cells Adenosine triphosphate (ATP) structure, showing the high-energy phosphate bonds. NH2 N N H N O HO P OH O O P OH O O P O CH2 O OH H H H H OH OH N H Figure 5-1. Typical behavior of single substrate-enzyme reactions. [Based on Baily and Ollis] (a) (b) 0.25 0.35 rmax = k*Et S = 1.5 0.25 r (mmols/L/s) r (mmol/L/s) 0.30 S = 1.0 0.20 0.15 0.10 S = 0.5 mmol/L 0.05 0.00 0.20 0.15 0.10 r = r max/2 0.05 S = Km 0.00 0.0 0.2 0.4 Et (mmol/L) 0.6 0.0 1.0 2.0 S (mmols/L) 3.0 Figure 5-2. Substrate activation and inhibition. r (mmol/L/s) 0.12 0.10 0.08 Inhibition 0.06 0.04 Activation 0.02 0.00 0.0 1.0 2.0 S (mmol/L) 3.0 4.0 Figure 5-3. Effect of pH on enzyme activity for various ranges of pKa. 1.2 pK: 4 - 10 [E] / [E]t 1.0 5-9 0.8 6-8 0.6 6.75 - 7.25 0.4 0.2 0.0 1 2 3 4 5 6 7 pH 8 9 10 11 12 13 Figure 5-4. Effect of temperature on hydrogen peroxide decomposition by catalase. (a) E = 3.5 kcal/mol, ΔHd = 55.5 kcal/mol, ΔSd = 168 kcal/(mol-K), β = 258 mm3/min. (b) Exponential fitted to equation in (a) at 20 and 25 deg C, r20 = 185.8 mm3/min, θ = 1.024 . [Based on Baily and Ollis] rmax k [E]t T exp( E / RT) r r20 1 exp( S d / R ) exp( H d / RT) 400 (b) 350 r (cu mm/min) 300 250 (a) 200 150 100 50 0 0 20 40 T (deg C) 60 80 T 20 Glycogen or starch All NADH2 and FADH2 produced go to the CYTOCHROME SYSTEM Glycolysis Glucose (C-6) Figure 5-5. 2 ATP CO2 (Nine steps) Glycolysis, fermentation, and the Krebs cycle. Inputs and outputs from each cycle are shown, as well as connections with metabolism of other biochemical compounds. Transition to Krebs Cycle 4 ATP 2 NADH2 Each Pyruvate x 2 (C-3) Acetaldehyde (C-2) pyruvate Each Fermentation pyruvate NADH2 NADH2 CoA-SH Lactic Acid (C-3) Fats Acetyl-CoA Oxaloacetic Acid (C-4) NADH2 Citric Acid (C-6) Malic Acid Amino Acids Isocitric acid KREBS CYCLE Fumaric acid CO2 FADH2 Succinic acid -Ketoglutaric acid (C-5) Succinyl-CoA (C-4) ATP GTP CO2 NADH2 NADH2 Figure 5-6. Mitochondrion structure, showing membrane structure, division into inner and outer compartments (e.g. Smith et al) Figure 5-7. The cytochrome electron transport system, with relationship to glycolysis and the Krebs cycle (based on Vander, et al and Martini, et al). Cytoplasm Glycolysis Pyruvate O2 Glucose ATP H2 O Outer mitochondrial membrane CO2 Inner mitochondrial membrane ADP +Pi Outer compartment Inner compartment FAD H2 O Krebs Cycle NAD NADH2 FMN 2e- 2H+ 2H+ FADH2 2H+ CoQ H+ 2e- Cyt b Cyt c 2H+ 2H+ Cyt 2ea3 Cyt a 2H+ ATP synthase Figure 5-8. Structure of a chloroplast. [Schaum’s Biology] Figure 5-9. Photosynthetic apparatus. [Schaum’s Biology] Figure 5-10. Structure of chlorophyll a and chlorophyll b. R O N Chlorophyll a : R = CH3 N O Mg N O N O O Chlorophyll b : R = CHO Figure 5-11. Absorption spectrum of chlorophyll and of a green leaf, and action spectrum for rate of photosynthesis versus wavelength. [Baily and Ollis] Standard electrode potential, mV Figure 5-12. Noncyclic photophosphorylation. -800 -600 -400 -200 0 200 400 600 800 1000 1200 1400 1600 NADPH P430 NADP 2e- Cyclic photophosphorylation ATP ADP P700 Z Cyt b563 Photosystem I 1/2O2 2eH2O P680 Two photons Two photons Photosystem II 2H+ 12ATP 12 Molecules of 3-Phosphoglycerate (C-3) 6 Molecules of CO2 (Photorespiration) 12ADP The Calvin Cycle O2 12 Molecules of 1,3-diphosphoglycerate (C-3) 6 Molecules of Ribulose 1,5-diphosphate (C-5) 12 NADPH Figure 5-13. CO2 fixation. 10 molecules PGAL 12ADP [Based on Raven] 12 Molecules of Glyceraldehyde 3-phosphate (PGAL) (C-3) 12ATP 2 molecules PGAL Carbohydrate Synthesis (glucose, sucrose, starch) 12 NADP