Warburg O
advertisement

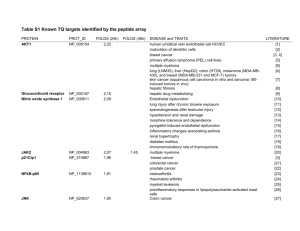
Otto Heinrich Warburg (Friburgo in Brisgovia, 8 ottobre 1883 – Berlino, 1 agosto 1970) Warburg O, Posener K, Negelein E. Uber den Stoffwechsel der Tumoren [On metabolism of tumors]. Biochem Z 1924; 152:319–344. Warburg O. The metabolism of London; 1930. tumours. Constable: Warburg O. On the origin of cancer cells. Science 1956; 123:309–314. Warburg O. On respiratory impairment in cancer cells. Science 1956; 124: 269–270. 90 years after Otto Warburg’s discovery we still ask ourselves “why do cancers have high aerobic glycolysis?” This question, however, can be understood in two different ways: 1. What is the cause of increased aerobic glycolysis in tumor cells? or 2. What is the advantage of increased aerobic glycolysis for tumor cells? Otto Warburg Nobel Prize in Physiology or Medicine 1931. Every year, between 20 and 25 Nobel Laureates spend a week in the Lake Constance area to meet the next generation of leading scientists. The Prime Cause and Prevention of Cancer Lecture at the meeting of the Nobel-Laureates on June 30, 1966 at Lindau, Lake Constance, Germany by Otto Warburg Director, Max Planck-Institute for Cell Physiology, Berlin-Dahlem Cell metabolism Quiescent versus proliferating cells Proliferating versus cancer cells The example illustrates that the biosynthesis of many cellular building blocks requires nutrients in excess of those needed for ATP production alone BIOMASS Glicolisi: Fase Esoergonica Decima reazione: seconda fosforilazione a livello del substrato _ O O C _ A D P O O A T P C C O C H 2 p ir u v a t o k in a s i PEP Il prodotto finale della 10° reazione è dunque il piruvato in forma chetonica. C O H C H 2 _ O O C O C t a u t o m e r ia C H 3 c h e t o e n o lic a Interconversione spontanea chetopiruvato enolpiruvato forma chetonica) (forma enolica) Pyruvate kinase isoforms • Liver (L) PKL gene • Red blood cell (R) • M1, expressed in most adult tissues PKM gene • M2, expressed predominantly in embryonic tissue and tumors The specific activity of PKM2 that is fully activated by FBP is approximately half that of PKM1. In the absence of FBP, PKM2 had less than one quarter of the activity of PKM1. Science. 2010 September 17; 329(5998): 1492–1499. Evidence for an alternative glycolytic pathway in rapidly proliferating cells Matthew G. Vander Heiden1,2,3,*, Jason W. Locasale2,3, Kenneth D. Swanson2, Hadar Sharfi2, Greg J. Heffron4, Daniel Amador-Noguez5, Heather R. Christofk2, Gerhard Wagner4, Joshua D. Rabinowitz5, John M. Asara2, and Lewis C. Cantley2,3,† 1Dana Farber Cancer Institute, Harvard Medical School, Boston, MA 02115 2Beth Israel Deaconess Medical Center, Division of Signal Transduction and Department of Medicine, Harvard Medical School, Boston, MA 02115 3Department of Systems Biology, Harvard Medical School, Boston, MA 02115 4Department of Biological Chemistry and Molecular Pharmacology; Harvard Medical School, Boston, MA 02115 5Lewis-Sigler Institute for Integrative Genomics and Department of Chemistry, Princeton University, Princeton, NJ 08544 Cancer Cell. 2012 November 13; 22(5): 585–600. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth Taro Hitosugi et al. 3-PG 2-PG BIOSINTESI DELLE PIRIMIDINE Ciclo di Krebs: enzimi O acetilCoA S C H3C CoA CoA H2 O COOH C O HO citrato - NADH+H + H C NAD+ COOH 8 COOH 2 2 2°deidrogenazione isomerizzazione H C HOOC C idratazione 3 CH 4 H FADH NAD NADH+H COOH CH2 GTP GDP+Pi CH2 COOH succinato 5 C S CoA + O CH2 CH2 chetoglutarato CoA SH CH2 COOH succinil-CoA succinato deidrogenasi COOH 4 O CH2 CoA SH + 6 CO 72 + C fosforilazione a livello del substrato FAD succinil-CoA sintetasi COOH 5 2 5 8 2°decarbossilazione ossidativa CO2 deidrogenasi 3 4 1°deidrogenazione COOH NAD + NADH+H 6 CH 6 OH isocitrato CH2 COOH 1°decarbossilazione ossidativa isocitrato deidrogenasi -chetoglutarato COOH 7 COOH fumarato 3 CH2 1 COOH malato 7 C 2 condensazione OH CH2 H2 O aconitasi CH2 1 ossaloacetato COOH COOH 1 COOH CH2 8 citrato sintasi SH fumarasi malato deidrogenasi glutaminolysis l’enzima malico catalizza la reazione che concorre a fornire equivalenti riducenti per la • sintesi degli acidi grassi + + C O O HN N A D A D H + H C O O H C H 2 C H 2 C H 3 C O H H C O C O O H + + N A D P N A D P H + H m a l a t o d e i d r o g e n a s i o s s a l a c e t a t o C O O H m a l a t o C O+ C O 2 e n z i m a m a l i c o C O O H p i r u v a t o Fase delle decarbossilazioni ossidative (reazioni 3, 4): AA 07/08 Reazione n°3: 1a decarbossilazione ossidativa. C O O H + + N A DN A D H + H H HC O C O O H CO CO C H 2 HC C O O H HC C O O H C H 2 C O O H is o c itr a to d e id r o g e n a s i C H 2 C O O H C O O H is o c it r a t o ( o s s a lo s u c c in a t o ) o s s id a z io n e C O 2 C H 2 C O O H c h e t o g lu t a r a t o d e c a r b o s s ila z io n e La reazione, catalizzata dall’enzima isocitrato deidrogenasi, prevede l’ossidazione dell’isocitrato con formazione di un intermedio, l’ossalsuccinato, dal quale si distacca una molecola di CO2. Si ha dunque formazione di NAD ridotto e liberazione di una molecola di anidride carbonica. L’ossalsuccinato viene decarbossilato mentre si trova legato all’enzima e non compare mai in forma libera glutamine glucose Acetyl-CoA from other sources (fatty acids, amino acids) % of total NADH/FADH2 production 60 30 10 Whatever its function, the occurrence of the Warburg effect reflects the activation of oncogenic signaling pathways whose physiological function is to promote glucose uptake and anabolic metabolism. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate Patrick S. Ward1,2 and Craig B. Thompson1,* 1 Cancer Biology and Genetics Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10065 2 Cell and Molecular Biology Graduate Group, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104 Cancer Cell. 2012 March 20; 21(3): 297–308. Via della serina GLICOLISI glucosio glutamina GLS-2 PHGDH PSAT piruvato Lattato o Acetil-CoA PSPH PHGDH: fosfoglicerato deidrogenasi PSAT: fosfoserina aminotransferasi PSPH: fosfoserina fosfoidrolasi GLS-2: glutaminase-2 Nat Genet. 2011 Jul 31;43(9):869-74. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Locasale JW, et al. Department of Systems Biology, Harvard Medical School, Boston, Massachusetts, USA Suppression of PHGDH in MDA-MB-468 cells caused a large reduction in the levels of a-keto-glutarate. In fact, of the major metabolites measured, aKG was the one with the most significant and largest change upon PHGDH suppression, whereas serine levels were not significantly changed. glutamine Science. 2012 May 25;336(6084):1040-4. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Broad Institute, Cambridge, MA 02142, USA. We measured the consumption and release (CORE) profiles of 219 metabolites from media across the NCI-60 cancer cell lines, and integrated these data with a preexisting atlas of gene expression. This analysis identified glycine consumption and expression of the mitochondrial glycine biosynthetic pathway as strongly correlated with rates of proliferation across cancer cells. Antagonizing glycine uptake and its mitochondrial biosynthesis preferentially impaired rapidly proliferating cells. Moreover, higher expression of this pathway was associated with greater mortality in breast cancer patients. Increased reliance on glycine may represent a metabolic vulnerability for selectively targeting rapid cancer cell proliferation. Original Article Recurring Mutations Found by Sequencing an Acute Myeloid Leukemia Genome Elaine R. Mardis, Ph.D., Li Ding, Ph.D., David J. Dooling, Ph.D., David E. Larson, Ph.D., Michael D. McLellan, B.S., Ken Chen, Ph.D., Daniel C. Koboldt, M.S., Robert S. Fulton, M.S., Kim D. Delehaunty, B.A., Sean D. McGrath, M.S., Lucinda A. Fulton, M.S., Devin P. Locke, Ph.D., Vincent J. Magrini, Ph.D., Rachel M. Abbott, B.S., Tammi L. Vickery, B.S., Jerry S. Reed, M.S., Jody S. Robinson, M.S., Todd Wylie, B.S., Scott M. Smith, Lynn Carmichael, B.S., James M. Eldred, Christopher C. Harris, B.S., Jason Walker, B.A., B.S., Joshua B. Peck, M.B.A., Feiyu Du, M.S., Adam F. Dukes, B.A., Gabriel E. Sanderson, B.S., Anthony M. Brummett, Eric Clark, Joshua F. McMichael, B.S., Rick J. Meyer, M.S., Jonathan K. Schindler, B.S., B.A., Craig S. Pohl, M.S., John W. Wallis, Ph.D., Xiaoqi Shi, M.S., Ling Lin, M.S., Heather Schmidt, B.S., Yuzhu Tang, M.D., Carrie Haipek, M.S., Madeline E. Wiechert, M.S., Jolynda V. Ivy, M.B.A., Joelle Kalicki, B.S., Glendoria Elliott, Rhonda E. Ries, M.A., Jacqueline E. Payton, M.D., Ph.D., Peter Westervelt, M.D., Ph.D., Michael H. Tomasson, M.D., Mark A. Watson, M.D., Ph.D., Jack Baty, B.A., Sharon Heath, William D. Shannon, Ph.D., Rakesh Nagarajan, M.D., Ph.D., Daniel C. Link, M.D., Matthew J. Walter, M.D., Timothy A. Graubert, M.D., John F. DiPersio, M.D., Ph.D., Richard K. Wilson, Ph.D., and Timothy J. Ley, M.D. Vol 361(11):1058-1066, September 10, 2009 Study Overview • A comparison of the genomic sequence of a tumor sample from a patient with acute myeloid leukemia (AML) and that of a normal skin sample from the same patient revealed an estimated 750 somatic mutations, of which 12 were in the coding sequences of genes and 52 were in conserved regions or regions with regulatory potential • Four mutations were found to be recurrent in AML, including mutations in NRAS, NPM1, IDH1, and a conserved region on chromosome 10 AML patients with the IDH1 R132 mutation harbored high serum levels of 2-HG. Maxmen Journal of Experimental Medicine 2010:0:jem.2072iti5-jem.2072iti5 © 2010 Maxmen In cytogenetically normal AML, mutated IDH1 is found in 10.9% of patients, while mutated IDH2 is found in 12.1%. Mutations in IDH1 are almost exclusively found at amino acid arginine 132 and IDH2 mutations are found more frequently at position R140 and less frequently at position R172. TET Cancer Cell. 2011 Jan 18;19(1):17-30. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. State Key Laboratory of Genetic Engineering, School of Life Sciences, Shanghai Medical School, Fudan University, Shanghai 20032, China. IDH1 and IDH2 mutations occur frequently in gliomas and acute myeloid leukemia, leading to simultaneous loss and gain of activities in the production of α-ketoglutarate (α-KG) and 2-hydroxyglutarate (2-HG), respectively. Here we demonstrate that 2-HG is a competitive inhibitor of multiple α-KG-dependent dioxygenases, including histone demethylases and the TET family of 5-methlycytosine (5mC) hydroxylases. 2-HG occupies the same space as α-KG does in the active site of histone demethylases. Ectopic expression of tumor-derived IDH1 and IDH2 mutants inhibits histone demethylation and 5mC hydroxylation. In glioma, IDH1 mutations are associated with increased histone methylation and decreased 5-hydroxylmethylcytosine (5hmC). Hence, tumor-derived IDH1 and IDH2 mutations reduce α-KG and accumulate an α-KG antagonist, 2-HG, leading to genome-wide histone and DNA methylation alterations. A novel inhibitor of mutant IDH1 inhibits human AML cell growth As inhibition of several signalling pathways did not selectively inhibit IDH1mut cells, we performed a computational drug screen using the ZINC library and the published crystal structure of mutant IDH1. By computational screening we identified a potential inhibitor of mutant IDH1 termed 2-[2-[3-(4-fluorophenyl)pyrrolidin-1-yl]ethyl]-1,4dimethylpiperazine (here termed HMS-101). Computational modeling showed that HMS-101 binds to the isocitrate-binding pocket of mutant IDH1. The IC50 for HMS-101 was significantly lower in mouse bone marrow cells transduced with IDH1mut compared to IDH1wt (1 μM vs. 12 μM, respectively, P<.001). Treatment of HoxA9+IDH1mut cells with HMS-101 at 10 μM significantly reduced intracellular R2HG levels in vitro. HMS-101 induced apoptosis in IDH1mut cells as evidenced by Annexin V staining and cell cycle analysis by BrDU