Replication, Transcription and Translation

DNA – The Molecule
Deoxyribonucleic Acid
Deoxyribonucleic Acid
• DNA is made of a phosphate group, a simple sugar and a nitrogenous base.
• Nitrogen Base- Organic Ring structure that contains one or more Nitrogen atoms
• Four Nitrogen Bases of DNA
– Adenine (A)
– Guanine (G)
– Cytosine (C)
– Thymine (T)
Note: No Oxygen on the #2 Carbon
Guanine- double ring structure
1
2
Adenine
Cytosine – single ring structure
Base Pairings
• Adenine pairs with Thymine
• Cytosine pairs with Guanine
• Adenine and Guanine are double ring structures called purines
• Cytosine and Thymine are single ring structures called pyrimadines
Ribonucleic Acid - RNA
• RNA is similar in structure to DNA.
• RNA is single stranded as opposed to double stranded (DNA)
• RNA has a ribose sugar as opposed to a deoxyribose sugar (DNA) see figure.
• The base pairs are A,C,G,and U
– Uracil replaces Thymine.
The Double Helix
• The spiral shape of DNA is called a double helix. It basically resembles a twisted ladder.
• The uniqueness of a strand of DNA is due to the order of the base pairs.
• Every time a cell reproduces, it must make an exact copy of its DNA.
Replication
• The process in which DNA is copied.
• Replication must be perfect, the method of replication is described as being semiconservative.
• semi-conservative- each new strand of DNA is composed of half original and half new material.
• Opposed to having one completely new strand of DNA and one completely original strand of
DNA
Chargaff ’ s Rule
• The amount of purines = the amount of pyrimadines.
• Therefore, in each species the amount of A=T and the amount of C=G.
• The amount of A,T, C and G in DNA varies from species to species.
• Both of these aspects of Chargaff ’ s rule helped to determine and understand the process of replication
DNA Replication-the process
• Replication is similar to unzipping a zipper.
• An enzyme breaks the hydrogen bonds between the base pairs.
• Then as the molecule unzips, free nucleotides pair up with each of the strands.
Steps in Replication
• 1. Unwinding- The old strand of parent DNA is unwound and unzipped.
– Helicase- enzyme that unwinds DNA.
• 2. Complementary base pairing- A pair with T and C with G.
– An RNA primer is added to initiate the process.
– DNA polymerase adds the base pairs, and removes the RNA primer.
Replication (continued)
• 3. Proofreading the strand- Most mistakes in pairings, and even some insertion of the wrong base pair or a deletion are corrected in the final step.
– Polymerase is the enzyme that is involved in this process.
• 4. The phosphate groups attach to the next ribose group with the help of the enzyme Ligase.
It ’ s not THAT simple
• During the replication process, nucleotides are joined in a 5 ’ to 3 ’ direction. (See drawing)
• Only one strand of DNA runs in a
5 ’ to3 ’ direction.
• In order for replication to occur quickly both strands must be copied at once.
• This is accomplished by having a leading and lagging strand.
Leading Strand
• As the molecule unzips the new DNA is synthesized going in the 5 ’ to 3 ’ direction heading toward the replication fork.
Lagging Strand
• The second strand works slower. It also must go in the 5-3 direction, but it goes away from the replication fork. Thus, leaving gaps of unpaired nucleotides.
• Okazaki Fragments- Pieces of replicated
DNA found on the lagging strand.
Responsible for speeding up the rate of replication.
DNA Replication- the figure
Polymerase Chain Reaction (PCR)
• Technique used to produce several identical copies of DNA using the enzyme polymerase.
• See Seven Daughters of Eve
Variability in DNA
• DNA can vary as a result of mutations and
“ jumping genes ” called transposons.
• Transposons- moveable genetic elements.
These are sections that control or suppress the expression of information.
• Because transposons have the ability to move from one part of the chromosome to another they can extremely alter the phenotype.
Transposons cause:
• Localized mutations. These are mutations that may occur in some cells but not others.
• Are a source of chromosomal mutations.
translocation, deletion and inversions
• Can leave a copy of themselves before they jump. duplication.
• Have been shown to be the basis of bacterial resistance to antibiotics.
Variability (cont.)- MUTATION
• Mutations in the base pair sequence
– Mutagen- environmental substance that causes mutations.
• Types of mutations
– Frameshift mutations- An insertion or deletion of a nucleotide which results in the changing of the genetic code.
– Point mutation- Change in a specific nucleotide which alters the codon. See page
247 for info on mutations and sickle cell anemia.
Chapter 16- Gene expression
• We have already discussed similarities/ differences of DNA and RNA.
• What is the location and function of DNA?
Location: nucleus, function: “ directions ” for protein synthesis
• What is the location and function of the ribosomes?
Location: cytoplasm, function: protein production
• Does anyone see a problem?
Directions for the product are NOT at the factory!
The Messenger
• We must have a messenger to carry the
“ info ” of DNA to the factory ” (ribosome)
Transcription Translation
• DNA mRNA ribo.
protein
The Central Dogma of
Molecular Biology
• The sequence of nucleotides in DNA codes for a sequence of nucleotides in RNA which directs the order of Amino Acids in a polypeptide (protein).
The players
• First the three types of RNA
– Messenger RNA (mRNA)- takes a message from DNA in the nucleus to the ribosome in the cytoplasm.
– Ribosomal RNA (rRNA)- along with some proteins, makes up the ribosome which synthesizes polypeptides (proteins).
– Transfer RNA (tRNA)- brings Amino Acids to the ribosome.
The processes- The Game don ’ t be a hater
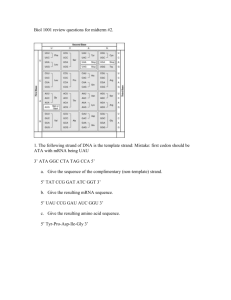
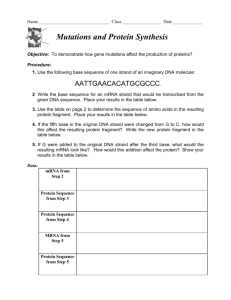
• Transcription- Process where a DNA strand serves as a template for the information on mRNA.
• Translation- Process where the sequence of codons in mRNA determines the sequence of Amino Acids in a polypeptide.
– Codon- three nucleotides in DNA or RNA which codes for a particular amino acid or termination of translation. See fig 17.4 pg 329
Transcription
• DNA TAC TGC CTG GCC ACT
• When rewritten as RNA
• RNA AUG ACG GAC CGG UGA
The Code: the language
Translation-decoding the RNA
• AUGACGGACCGGUGA
• AUG- methionine
• ACG- threonine
• GAC- aspartate
• CGG- arginine
• UGA- stop
Transcription- the details
• Transcription is started by the promoter region.
• When RNA is produced it is often shorter than the template strand of DNA.
• mRNA is processed and so that only particular sections of DNA end up in the mature segment of mRNA.
Transcription
• Only one strand of DNA is the template for mRNA production. It is called the template strand.
• The template strand contains the promoter region. There can be multiple promoter regions on one strand. As a result, RNA production can occur at multiple sites on a
DNA strand.
• The strand that is not used for mRNA production is called the inactive strand.
The Processing of mRNA
Introns-Portions of primary mRNA which are removed from mRNA.
Introns can be thought of as DNA which does not bind to RNA. Whatever proteins are coded for by the bases on these sections of DNA will NOT be produced.
The presence of introns suggests that there is more DNA than is necessary.
The role of introns is still under investigation.
Exons- Portions of DNA which bind to
RNA. Whatever traits these segments code for will be expressed.
mRNA
• mRNA that still contains introns is called primary mRNA.
• A complex of special protiens called spliceosomes remove the introns, leaving behind just the exons on a strand of mRNA.
• The mature mRNA leaves the nucleus through a pore in the nuclear membrane.
Translation requires three steps
• 1. Initiation- A small ribosomal unit attaches to the start codon (AUG). The first tRNA called the initiator tRNA pairs with the codon. Then a large ribosomal unit attaches and translation begins.
– Anticodon- A group of three bases on the tRNA that is the compliment to the three bases of the codon on the strand of mature mRNA.
Structure of tRNA- note the anticodon
Translation step 2: Chain elongation
• Amino acids are added in a long chain to form a peptide.
• Elongation occurs when the tRNA moves from one site on the ribosome to another. (see figure
16.10).
• The amino acids brought in by previous tRNA is added to the current tRNA. Thus extending the chain of amino acids in a specific order as specified by the mRNA.
Chain elongation
Step 3. Chain termination
• The stop codon does not code for a particular amino acid and causes peptide production to cease.
• The tRNA is cleaved and the peptide leaves the ribosome.