Mass Action:
advertisement

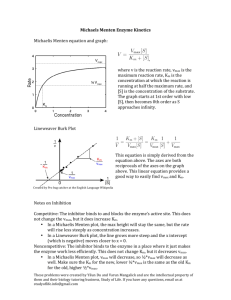
Today we will deal with two important Problems: 1. Law of Mass Action 2. Michaelis Menten problem. Creating Biomodel in Vcell we will solve these two problems The Law of Mass Action (by Norwegian scientists 1864–79CCato M. Gulberg and Peter Waage) Cosider this chemical reaction equation in which reactants A and B react to give product AB. A + B AB The mass action law states that if the system is at equilibrium at a given temperature, then the following ratio is a constant. [ AB] K eq [ A][ B] That is, the rate of a reaction is proportional to the product of the active masses of the reagents involved. This is the ideal law of chemical equilibrium or law of mass action. More explicitly.... In a chemical reaction --kr A+B AB kf the "chemical affinity" or "reaction force" between A and B did not just depend on the chemical nature of the reactants, but also depended on the amount of each reactant in a reaction mixture. The affinity or the reaction force between A and B = kf [A] [B] Kf is affinity constant. forward reaction rate= kf [A] [B] backward reaction rate= Kr[AB] At equillibrium forward reaction rate = backward reaction rate Kf [ AB] K eq Kr [ A][ B] Now create a New Biomodel Document Consider the case where A and B reacts to produce AB A+BAB. Lets start with the following steps--- •Give name of the unnamed compartment •Add specieses A,B and AB •Set reaction •Set Reaction Kinetic Editor •Kinetic type Mass Action •Put Value of Kf and Kr •Save the Model with a name. •Go to Application NewDeterministic •Name the Application (again we will be doing compartmental application) and OK. •Put volume=1 in Structure Mapping Text. •Click Initial Condition Text and assign Value of A and B. •Save the Model once again to View Math and to start Simulation. Note the Mathcomment,Which says Math Model is already generated by the software. We can view Math Equations or View VCMDL Now click simulation Text to run the Simulation. See Results and play with the different parameters by clicking Edit . Exercise: Run Simulation with the following parameter value---Kf =1 , Kr =1, Ainit= 1, Binit = 1 Check these two points. - Keep all parameters the same as given on this slide, and start increasing parameter Ainit. Make initial AB zero. Is equilibrium reached faster? Does stable AB concentration increase? Why? - Keep all parameters the same as given on this slide, and start increasing parameter kr. Make initial AB zero. Is equilibrium reached faster? Does stable AB concentration increase? Why? Keep all the parameters =1.0 and increase Ainit, see the stable concentration of AB. Increase Kr, see Equillibrium is reached faster. At equillibrium, forward reaction rate = backward reaction rate Go back to the equations to verify your results. Michaelis-Menten Kinetics(by Leonor Michaelis and Maud Menten in 1913 ) M-M kinetics approximately describes the kinetics of the Enzyme. The most convenient derivation of the Michaelis–Menten equation, is obtained as follows: The enzymatic reaction is assumed to be irreversible, and the product does not bind to the enzyme. Kf Kp E+S ES E+P Kr Enz+SubEnz-Subs-complex Enz+Prod Now our aim is to analyze this mechanism. With Vcell we can find the reaction rate of production of the product P and the complex ES and also study the impacts of several rate constants Kf,Kr,Kp. Now, consider the rate of effective production of P : dP dt The Michaelis-Menten equation relates the reaction rate V = to the substrate concentration [S]. The corresponding graph is a hyperbolic function; the maximum rate is described as Vmax. Vmax [ S ] dP V K M [S ] dt KM Kr Kf ,Vmax =KP* [E]in Start with a new Biomodel: •Name the compartment •Add Specieses S, E, ES, P •Set the Reactions •Set Reaction kinetic Editor for two reactions. •Set General as the kinetic Type Now we have to set the Reaction Rates. Here We have two reactions: 1.Substrate binding, where E and S react to produce the complex ES Reaction rate J_reaction0= Kf*[E]*[S] - Kr*[ES] - KP*[ES] 2.Catalytic Step, where ES dissociates to produce P and E Raction rate J_reaction1 = KP*[ES] Now go to File>Save as... to save the model. Remember two assumptions: 1. A lot of substrate molecules, very few ‘expensive’ enzymes ( [S]>>[E]), so [S] does not change much for a long time. 2. The system is in steady-state, i.e. that the ES complex is being formed and broken down at the same rate, so that overall [ES] is constant . Steps are same as Mass-Action •Go to ApplicationNewDeterministic •Name the Application and OK. •Put volume=1 in Structure Mapping Text. •Click Initial Condition Text and assign Value of E and S.(Remember E<< S) •Save the Model once again to View Math and to start Simulation. •Math Model will be generated Automatically. Clicking View Math we can see the Math Model generated by the software With the radio button we can see the value or name of the parameter eqn. Run the Simulation: Clicking Edit we can change different parameters •Analyze formation rate of ES, P with time or with other parameter. •We can choose Graphical results or data values by clicking Exercise: Consider the parameter values as follows: Kf 1 1 1 , Kr , Kp , E in 1 M , S in 100 M , ES in 0, P in 0 M sec sec sec 1. Run simulation for t= 1, 5 and 10 seconds. 2. Keep all parameters the same as given on this slide. Solve the chemical reaction equations. Discuss the results. Compute V, check if the formula for V in the previous slide is correct. 3. Make S in 1 M and solve the chemical reaction equations again. Discuss the results. Does Michaelis-Menten approximation still work? Vmax [ S ] dP V K M [S ] dt ,Vmax =KP* [E]in KM Kr Kf See math description for J_reaction1 J_reaction1 = V, calculate V. Check how V changes with [S]in For small value of S, say [S]in = 1.0 uM