Electrochemical Studies of our Beloved Nitrophorins

Rhodnius Nitrophorins:
Binding Constant and
Electrochemcial Measurements
Max Shokhirev
University of Arizona
Chemistry Department
Walker Lab
The kissing bug…
• Nitrophorins are found in the salivary glands of
Rhodnius Prolixus which is commonly known as the kissing bug.
•
Rhodnius is native to the Amazon River Delta and has spread as far north as Arizona and Texas
•
Rhodnius also helps spread Chagas disease throughout
Central and South America, which can be fatal.
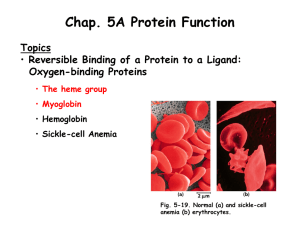
Nitrophorins
• Belong to the lipocalin family.
• A Histidine residue coordinates the heme
• Bind NO and often other ligands
• Nitrophorins 1-4 found in the saliva of fifth instar
Rhodnius. Nitrophorin 5 and 6 found only during the first instar. NP7 has been expressed in a laboratory setting, but hasn’t been identified in the bug itself 1
Nitrophorin Environment
Yummy
Blood!
Salivary glands: pH ~5.5; NO bound
NO displaced by Hm, which is present at a higher concentration at the site of the bite
Host tissues: pH 7.3; Hm bound
General Nitrophorin Function
• Nitrophorins are
N O transport proteins
• Use a heme cofactor to bind and release the
NO
• The Fe(III) heme is stabilized in order to prevent auto reduction of the Fe center
– NO binding to Fe(II) is irreversible!
• NO released in the host tissues causing vasodilation.
Other Nitrophorin Functions
• Anticoagulant properties of NP2 and NP3
– NP2 was found to bind to factor IX of the Xase coagulation system 2 .
• Platelet aggregation inhibition by NP7
– Through an NO mediated mechanism 3
• Histamine binding serves an anti-inflammatory function
Nitrophorin Similarity
• Nitrophorins 1-4 share
38% sequence identity
• NP2 and NP3 have
79% sequence identity
• NP1 and NP4 have
90% sequence identity
• All share the same structural motifs:
– Eight stranded beta barrel with a b-type heme within the barrel
Structure of NP2
GH Loop
EF Loop
Heme
Center
AB Loop
His 57
From PDB 1T68 4
One way to study a protein…
• Site-directed mutagenesis studies
– Synthesize NP2 with a select amino acid changed.
– Observe the changes in redox potential , ligand binding constants, NMR, EPR…
– Rationalize the observed trends.
– Publish a paper.
What I do…
• Mostly Spectroscopy
– Binding Constant Measurements
– Electrochemical Measurements
– A little Cyclic Voltometry
Binding Constant Measurements
• The UV-vis spectrum of nitrophorins changes upon ligand binding.
• The ratio of ligand-bound to ligand-free can be measured by observing the change in absorbance for one species as a function of changing ligand concentration
• We can fit the resulting data using the following tight ligand binding equation:
Example Measurement
0.3
0.25
0.2
0.15
0.1
0.05
325
NP2 D1A D89A pH5.5 Im binding
NP2 D1A D89A pH5.5 Im binding
0.16
0.15
0.14
0.13
0.12
0.11
0 2e-5
[Im]
4e-5
375 525 425 475
Wavelength (nm)
575
6e-5
Binding Constant Measurements
• This is a powerful method for the study of a protein such as a nitrophorin:
– Allows us to observe how pH affects ligand binding
– Allows us to compare different ligands with respect to nitrophorin affinity for those ligands
– Each mutant studied in this way reveals something about the importance of the mutated amino acid to protein function.
Electrochemistry
• NO tends to auto reduce Fe(III) to Fe(II) if the
Fe(III) is not protected.
– NO dissociation constants from Fe(II) in picomolar range
• Nitrophorins stabilize the Fe(III) form of the heme center through both heme ruffling and negatively charged residues near the heme center.
• It is possible to measure the potential at which half of the nitrophorins in solution are reduced by observing the spectrum of the nitrophorin solution at different applied potentials
Electrochemical Studies
• Measuring redox potential:
– Different pH values
– With ligands bound (NO, Hm, Im) or just H
2
O
• Redox potential related to concentration through the Nernst Equation:
Physical Setup
Spectrophotometer:
Records the spectrum of the protein in the electrochemical cell
Potentiostat:
Provides a steady measurable electrical potential
Physical Setup
Argon Gas:
Helps prevent oxygen contamination of the protein solution
The Electrochemical Cell
Auxiliary
Electrode
Argon Gas
Reference
Electrode
Working Gold electrode reduces the protein at the cell window.
Light
0.5
0.4
Electrochemical Data
0.48
0.47
0.46
0.45
0.44
0.43
0.42
0.41
0.40
Absorbance vs Applied Potential
-600 -500 -400 -300 -200 mV ( vs . silver electrode)
-100 0
0.3
400 500
Wavlength (nm)
600
The Electrochemical Cell
Thank you…
References
• 1) Moˆnica F. Moreira; Heloisa S.L. Coelho; Russolina B. Zingali; Pedro L.
Oliveira; Hatisaburo Masuda (2003) Insect Biochemistry and Molecular
Biology 33, 23-28.
•
2) Nanda P. Gudderra; Jose ´ M. C. Ribeiro; John F. Andersen. (2005) J.
Biological Chemistry 280 , 25023-28.
• 3) John F. Andersen; Nanda P. Gudderra; Ivo M. B. Francischetti; Jesus G.
Valenzuela; and Jose´ M. C. Ribeiro. (2004) Biochemistry 43 , 6987-96
• Ribeiro, J. M. C.; Hazzard, J. M. H.; Nussenzveig, R.; Champagne, D.;
Walker, F. A. (1993) Science 260 , 539-541.
• Shokhireva, T. Kh.; Berry, R. E.; Uno, E.; Balfour, C. A.; Zhang, H.; Walker, F.
A. (2003) Proc. Natl. Acad. Sci. USA 100, 3778-3783.
•
Andersen, J. F.; Montfort, W. R. (2000) J. Biol. Chem.
275 , 30496-30503.
•
Roberts, S. A.; Weichsel, A.; Qiu, Y.; Shelnutt, J. A.; Walker, F. A.; Montfort,
W. R. (2001) Biochemistry 40 , 11327-11337.
Acknowledgements:
Dr. Walker, Dr. Berry, Dr. Shokhireva, Honjun
Zhang, and the rest of the Walker Lab