a new UPOV model - Raad voor plantenrassen
advertisement

DNA profiles in DUS testing of grasses
A new UPOV model ?
Henk Bonthuis
Naktuinbouw
Aanvragersoverleg Rvp
Wageningsche Berg 19 oktober 2015
Lolium perenne (perennial ryegrass)
•
Genetically diverse:
–
–
–
–
•
Obligate outcrossing species
genetically heterogenic populations
Synthetic Varieties:
created by polycross of selected individual clones (3-20)
Morphologically diverse
– Relative uniformity (in relation to existing varieties)
•
Additional diversity:
– Genotype x location interactions
– Environmental effects (winterhardiness,drought, stress)
– Random experimental errors
Challenges
• Make DUS testing of Grasses more efficient
–
–
–
–
DUS testing of grasses is labour-intensive
testing based on single plants
measured characteristics mainly
large reference collection
• Make DUS testing of Grasses more predictable
–
–
–
–
Unpredictable morphological differences at the start of DUS
Therefore ref. collection needs to be measured completely each year
Low discriminative power due to uncontrolled environmental variation
Many negative DUS reports as a result
Pilot study (2014)
Objectives
• Making DUS testing of grasses more efficient by using
UPOV Model 2 approach:
– combining morphological and molecular distances
for the management of the reference collection.
• Making DUS testing of grasses more predictable by
creating molecular database(s):
– to be used by (all) Examination Offices
– to be used by breeders for DUS screening beforehand
Approved UPOV model 2 approach
Setting a Molecular threshold for reference varieties
to be excluded from the field trial
UPOV-BMT Model 2
Morphological distance
• Grasses today:
growing the full
reference
collection
Variety pairs
to be tested
in the field
0
Molecular distance
UPOV-BMT Model 2
Morphological distance
0
Molecular distance
•
Facts well known:
•
Morphological
threshold for
distinctness
•
Variety pairs
above morph.
threshold were
actually
redundant
UPOV-BMT Model 2
Morphological distance
• Additional
information from
molecular profile:
Molecular distance
of variety pairs
0
• Can varieties with
large molecular
distances be excluded
from the field trial ?
Molecular distance
UPOV-BMT Model 2
Morphological distance
•
Area of Concern
probability of incorrect
decisions on excluding
reference varieties
from the field trial
Area of
concern
0
Molecular distance
UPOV-BMT Model 2
Morphological distance
0
Molecular distance
•
Incorrect
decisions can
be avoided
•
by setting the
molecular
threshold at a
safe level for
morphological
distance.
Variety pairs to
be excluded from
the field trial
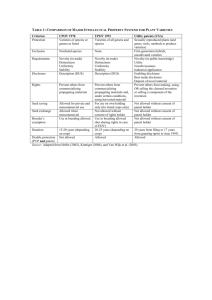
UPOV model 2 in Maize (France)
• Purple area = varieties which can be excluded from the field trial,
based on Rogers distance and morphological GAIA distance
UPOV model 2 in potato (NL)
Combining Morphological and Molecular distances
5 pairs not distinct:
Mutants and/or
closely related
varieties
0,7
0,6
Cityblock distance
0,5
0,4
0,3
Rest of 16653 pairs
were all distinct
0,2
0,1
0
0
0,2
0,4
0,6
0,8
1
Jaccard distance
•
Based on validated data of 183 varieties (16653 pairs)
Combining Morphological and Molecular distances
Thresholds for
distinctness:
0,7
0,6
Cityblock distance
0,5
Morphological distance:
Cityblock 0,05
0,4
0,3
Molecular distance:
Jaccard 0,15
0,2
0,1
0,05
0
0
0,2
0,4
0,6
0,8
1
Jaccard distance
•
Threshold for molecular distance based on 16,653 pairs (minus 5) corresponds with
threshold previously found in the SSR database project (900 varieties > 400,000 variety
pairs), confirmed by present database (1953 varieties = 1,906,128 variety pairs)
Combining Morphological and Molecular distances
Distinct Plus
More distinct than just distinct
0,7
0,6
Cityblock distance
0,5
Varieties which can be excluded
from the growing trial:
0,4
Cityblock distance > 0,10 and
Jaccard distance > 0,20
0,3
0,2
0,1
0,10
0,05
0
0
0,2
0,4
0,6
0,8
1
Jaccard distance
•
•
High-lighted area: above distinct plus thresholds
low risk for wrong decisions on reference varieties to be
excluded from the growing trial
Pilot study on Grasses
• Lolium perenne (perennial ryegrass)
• Phenotypical data of 20 amenity-type varieties
• 20 varieties make (20x19/2 =) 190 variety pairs
–
–
–
–
standard UPOV characteristics – TG/4/8
16 morphological traits
measurements of 60 individual plants per variety
Complete dataset over 3 years (2010 – 2012)
Trait summary & weights used in distance calculation
Trait description
min
mean
max
range
weight
Growth habit
4.5
5.1
5.5
1.0
1
Intensity of green colour
4.9
5.6
7.3
2.4
1
% flowering in autumn
1.6
1.7
2.4
0.8
1
Heading date
47
57.8
65
18
9
Flagleaf length (mm)
91.3
111.4
125.9
34.6
6
Flagleaf width (0.1 mm)
34.6
38.2
44.5
9.9
4
Flagleaf length/width ratio
24.2
29.2
32.7
8.5
1
Flagleaf area
3019.1
3737.4
4585.7
1566.6
1
Plant height 30 days after heading
58.6
63.4
70.4
11.8
6
Length upper internode
180.3
220.2
244.9
64.6
6
Inflorescence: length
137.2
148.4
166.7
29.5
1
Length of longest stem
323.5
372.7
411.5
88
6
Inflorescence: number of spikelets
18
19.7
20.7
2.7
6
Inflorescence density
7.1
7.7
8.7
1.6
1
Length outer glume (mm)
6.7
8.1
9.8
3.1
4
Length basal glume (mm)
10.6
11.6
13.3
2.7
4
Genotyping-by-Sequencing (GBS)
• 20 varieties of amenity grasses
– Genotyped by AgriBio lab (Centre for AgroBioscience,
Bundoora, Victoria, Australia)
– 1000 seeds/variety - representing variety (population)
– DNA extraction of bulk sample (DNeasy Plant kit from Qiagen)
– Profiles based on allele frequencies
– Targeted amplification step
– Ligation using bar-coded synthetic DNA adapters
– Sequencing with Illumina MiSeq
– 295 SNP-markers retained
Methods: calculating distances
• Distances between varieties based on morphological traits:
– Euclidean, Cityblock, Minkowski, Divergence, etc.
• Distances between varieties based on SNPs:
– Euclidean, Jaccard, Rogers, Nei, etc.
– ∑k { wk(xik, xjk) sk(xik, xjk) } / ∑k { wk(xik, xjk) }
•
•
•
•
Xik , Xjk = value of the data variate k in unit i or unit j resp.
Sk = contribution function (depending on the variate range)
Wk = weight function (1 for all QN-variates)
For further details see: Gower, 1971/1985
Data Analysis
• Calculated different distance measures for morphological traits
(Euclidean, Cityblock, etc) based on range and weights
• Calculated different distance measures for SNPs: Euclidean, Jaccard
• Considered combination of the two types of distances (UPOV-Model 2)
• Selected SNPs with higher correlation to morphological traits
• Selected 111 SNPs with a correlation >0.5 with a trait
Results: Genetic relationships
Varieties
genetically
sufficiently
distinct
(based on Nei’s
coëff for SNPs).
Nautica most
divergent.
Greenway and
Hayley most
similar.
(Trojan and
Nagano are
control varieties)
Combining Y: Molecular distance (Euclidean)
and X: Morphological distances (Cityblock) and Ndiff
(Number of trait differences) for 190 variety pairs
Molecular
threshold
27 pairs
GxE interaction for morphology
interfering with molecular threshold for distinctness
Molecular
threshold
27 pairs
Conclusions of Pilot (end 2014)
• UPOV Model 2 does not work for grasses
• Due to failing morphological model of Lolium perenne
• Morphology = limiting factor:
too many GxE interactions, environmental effects and
experimental errors involved.
Failing Morphological Model of grasses
• Varieties of perennial ryegrass should be distinct (by nature) !
– Obligate outcrossing species, genetically heterogenic populations
– Synthetic varieties created by polycross of selected individual clones
• Too many GxE interactions and environmental effects
– Observations on single plants, randomly picked leaves, seasonal effects, etc.
• O.P. Crops excluded from PBR failing to fulfill DUS criteria in 1960’s.
– Sugar beet, Rye, Alfalfa, White Clover, Caraway, etc. (ZPW 1967).
• Narrowing genepools in grasses (since 1960’s) ?
– Too much noise in relation to real genetic differences
– puts additional pressure on morphological model of grasses
New approach
presented by US experts from Monsanto at UPOV-BMT – Korea 2014
Candidates described in relation to Reference Varieties (based on molecular distance)
UPOV – BMT 2014
Molecular distances based on reference varieties
added to the morphological description as additional traits
Distance application to genotypes: Identify reference varieties:
by enlarging database – mapping (all) varieties in common knowledge
Example: data Pilot project
Phylogram illustrating
separate genepools:
tested EU cultivars –
amenity types (in blue) and
varieties from the Australian
perennial ryegrass catalogue
known at AgriBio Lab
(mostly fodder types).
(Control samples in red)
New challenge ahead
Estimate genetic variation
representative for morphology
excluding environmental influences
Ongoing efforts on Lolium perenne at Naktuinbouw
•
Expanding and Improving the set of SNPs for maximum differentiation
– Ongoing GBS project financed by Rvp (2015 and 2016) – but limited resources
– Create consortium of Labs, EO’s and breeders for maximum impact
•
Identification of reference varieties
– Reference varieties (i.e. additional traits) primarily needed for variety description
– Reference varieties should be relevant for the area under consideration
•
Define molecular thresholds for distinctness (crucial !)
–
–
–
–
–
–
–
Excluding environmental effects from morphological data
Requires genome-wide SNPs and Bio-informatics tools
Include datasets from different environments (estimating GE and e)
Associate phenotype and genotype by genomic prediction (training and target pop) ?
Calculate thresholds for distinctness (and distinct plus)
Molecular thresholds determine direct variety comparisons (target oriented testing)
Morphology remains ultimate test for distinctness
Ultimately …
• To make breeding more effective
• DUS testing of grasses should be
more efficient and more predictable
• Showcase for other (cross-pollinated) crops ?
• New UPOV-BMT model ?
Acknowledgements:
João Paulo (Biometris, Wageningen)
Paul Goedhart (Biometris, Wageningen)
Noel Cogan et al. (Biosciences Research, Bundoora,
Australia)
Quality in Horticulture