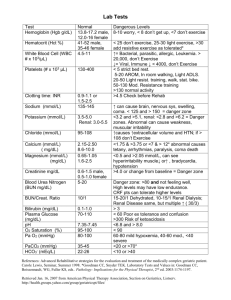
Online Supplemental Information
advertisement

Rosc-Schlüter et al. Online Supplement CVR-2010-1473.R2 ONLINE SUPPLEMENT NOX2-derived reactive oxygen species are crucial for CD29-induced pro-survival signaling in cardiomyocytes Berit I. Rosc-Schlütera, Stéphanie P. Häuselmanna, Vera Lorenza, Michika Mochizukia Federica Facciottib, Otmar Pfistera,c and Gabriela M. Kustera,c a Myocardial Research and b Experimental Immunology, Department of Biomedicine, University and University Hospital Basel, Switzerland and c Division of Cardiology, University Hospital Basel, Switzerland Short Title: Nox2 mediates CD29-induced survival signaling Correspondence: Gabriela M. Kuster, M.D., Myocardial Research, Department of Biomedicine, and Division of Cardiology, University Hospital Basel, Hebelstrasse 20, 4031 Basel, Switzerland Tel: ++41 61 328 77 36, Fax: ++41 61 265 23 50, Email: GKuster@uhbs.ch Rosc-Schlüter et al. Online Supplement CVR-2010-1473.R2 SUPPLEMENTAL MATERIAL Material for cell culture: reagents from GIBCO/Invitrogen: DMEM (low glucose, 1g/L); phenol red free (prf)-DMEM (low glucose, 1 g/L), HBSS, Hepes, 0.05% trypsin-EDTA, PBS, L-glutamine (GlutaMAX), penicillin (P, 100 U/mL), streptomycin (S, 100 µg/mL) and FCS; 5-bromo-2deoxyuridine (BrdU, 200 mmol/L) and trypan blue were purchased from Sigma Aldrich, collagenase from Worthington, accutase from PAA Laboratories (The cell culture company). Treatments: Wortmannin, U0126, apocynin, DPI (Diphenyleneiodonium chloride), PMA and H2O2 were purchased from Sigma-Aldrich; LY294002 [2-(4-Morpholinyl) 8-phenyl-4H-1-benzopyran-4one], Akti1/2 (Akt inhibitor VIII) and MnTMPyP [Mn(III)tatrakis(1-methyl-4-pyridyl) porphyrin pentachloride] were purchased from Merck. Activating anti-β1-integrin (α-CD29) antibody was from BD Biosciences, inhibitory α-CD29 from BioLegend, rabbit IgG from Sigma Aldrich and mouse IgG from Abcam. Viral transfection, siRNA: The following viral constructs were purchased from the Gene Transfer Vector Core, University of Iowa: Ad5CMVCatalase1, Ad5CMVSOD12 and Ad5CMVcytoLacZ3. Rat NOX2 siRNA (J-093524-05-0020) and control siRNA (non-targeting sequence) (D-001810-01-20) were purchased from Thermo, and the Amaxa Rat Neonatal Nucleofector kit from Lonza. Flow cytometry: 5-(and-6)-chloromethyl-2’,7’-DCF-DA (dichlorodihydrofluorescein diacetate acetyl ester, CM-H2DCFDA) was purchased from Invitrogen . Sample preparation: Cell lysis buffer (Cell Signaling Technology, Inc., Danvers, MA, USA): 10x; 20 mM Tris-HCl (pH 7.5), 150 mmol/L NaCl, 1 mmol/L Na2EDTA, 1 mmol/L EGTA, 1% Triton-X100, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerophosphate, 1 mmol/L Na3VO4, 1 μg/mL leupeptin; 1 mmol/L Pefabloc SC Plus (Roche Diagnostics GmbH, Mannheim, Germany) and phosphatase inhibitor cocktail 1 (Sigma-Aldrich). Sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western Blotting: DualColor Protein Loading Buffer Pack (including a reducing agent), Page RulerTM Prestained Protein Ladder from Fermentas were used. Anti-phospho-ERK1/2 [p44/42-MAPK, (Thr202/Tyr204)], antiphospho-MEK1/2 (Ser217/221), monoclonal anti-phospho-Akt [(Ser437) (193H12)], anti-phosphoGSK-3β (Ser9), and corresponding anti-total antibodies p44/42 MAP kinase, MEK1/2, Akt, monoclonal GSK-3β rabbit were from Cell Signaling. For adenoviral experiments, anti-Cu/ZnSOD and anti-catalase antibodies purchased from R&D were used to monitor expression of the respective target protein. Secondary antibodies [horseradish peroxidase (HRP)-conjugated AffiniPure goat αrabbit- and rabbit α-goat (for catalase) IgG] were purchased from Jackson Immuno Research Laboratories. PVDF-membrane (Polyvenylidene fluoride, HybondTM-P) was from GE Healthcare BioSciences. Western lightening plus (enhanced chemilumiscence, ECL) was from PerkinElmer. Blue XRay films from Thermo Fisher Scientific were used. Lucigenin Assay: Jude Krebs (JK) Buffer: 119 mmol/L NaCl, 20 mmol/L Hepes, 4.6 mmol/L KCl, 1 mmol/L MgSO4, 0.15 mmol/L Na2HPO4•2H2O, 136.09 mmol/L KH2PO4, 84.01 mmol/L NaHCO3, 147 mmol/L CaCl2, 180.2 mmol/L glucose. Rosc-Schlüter et al. Online Supplement CVR-2010-1473.R2 SUPPLEMENTAL METHODS Isolation of neonatal rat ventricular myocytes (NRVM). NRVM were isolated from 1-3 days old pup hearts as previously described 4, 5. Hearts were removed from Wistar (-12/2010) or Sprague-Dawley (2011-) rat pups sacrificed by decapitation. After ventricles were dissected from the hearts, they were washed in HBSS and cut in equal pieces. Ventricular pieces were digested in 0.05% trypsin-EDTA in a 50mL tube (Sarstedt) for 14 hours overnight with gentle agitation at 4°C. Trypsin-EDTA was removed and pieces were washed once with 7%-FCS DMEM by gentle shaking in a pre-warmed water bath at 37°C for 4 minutes. After removing the FCS containing DMEM, step-wise (4-7 minutes each) digestion with 0.07% (w/v) collagenase in 50 mL HBSS was performed. Aliquots were collected in 50ml tubes containing 10 ml 7% FCS-DMEM and kept on ice. After digestion was completed, the collected cell suspension was filtered with a 100 µm cell strainer to remove residual tissue parts and centrifuged for 8 minutes at 800 rpm and 4°C. Pellets were resuspended in 10 ml 7%-FCS-DMEM. Cell suspensions were pre-cultured at 37°C, 5% CO2 in T75 culture flasks (Sarstedt) for 2 hours to allow fibroblasts to attach, whereby after 1 hour of incubation the cell suspension was transferred to new T75 culture flasks. The cell number was estimated using a Neubauer hemocytometer counting chamber (Carl Roth) with trypan blue as an exclusion dye for dead cells. The yield of viable cells was estimated as follows: Mean (total number of cells in 4 grids/ 4) 2 104 (cell suspension volume). 2x106 cells were plated in 7% FCS-DMEM [with P/S, 25 mmol/L Hepes and 100 µmol/L BrdU (to prevent proliferation of non-myocytes)] on a 60 mm plastic dish (P60, BD Falcon). NRVM were kept for 24 hours at 37 °C and 5% CO2 before changing to serum-free (SF)-DMEM (without FCS, but with Hepes and P/S) for an additional 16 hours. Isolation of neonatal mouse ventricular myocytes (NMVM). NMVM were isolated from 1-2 days old pup hearts of wild-type (C57/BL6), p47phox LOF (homozygous Ncf1m1J) and NOX2 (gp91phox, homozygous Cybbtm1Din) knockout strains as described previously 6, 7. Collected ventricular heart pieces were digested in 0.05% trypsin-EDTA for 45 minutes with gentle agitation at 4°C, followed by a wash step with 10% FCS-DMEM, and serial digestions with 0.0686% (w/v) collagenase in HBSS. Non-myocytes were removed by pre-plating on T75 Sarstedt-flasks at 37 °C and 5% CO2 for 2 h. The cell number was estimated using a Neubauer hemocytometer counting chamber [yield= Mean (total number of cells in 4 grids/ 4) 104 (cell suspension volume)]. 10% FCS-DMEM (with P/S, Hepes and BrdU) was used to plate cells of each strain on laminin pre-coated (70 µg/mL) 35 mm dishes (P35) or 12-well plates. Cardiomyocytes were seeded at a density of 0.9x106 (12-well plate) or 1.8x106 (P35) and kept at 37°C and 5% CO2 for 24 h. The serum-containing medium was replaced by SFDMEM 16 to 18 hours prior to treatment. Culturing of H9c2 cardiomyoblasts. H9c2 cells were cultured in DMEM containing P/S and 10% FCS at 37°C in 5% CO2 8. Treatments. NRVM were treated with W, LY, Akti1/2, U, DPI (all in DMSO) or MnTMPyP (in water) 45 minutes or apocynin (500 µmol/L, in DMSO) for 16 to 18 hours prior to α-CD29 administration, and DMSO was also added to the respective control dishes. α-CD29 was administered for 15 minutes, except for time course experiments. Sample preparation. All signaling samples were washed twice with ice-cold PBS and scraped in 70 µl (P35) or 120 µl (P60) cell lysis buffer, centrifuged at 15’000 rpm for 15 min at 4 °C, and supernatants were collected in new micro-tubes (Eppendorf). Cell lysis buffer (1:10 in ddH2O) was supplemented with 1 mmol/L Pefabloc SC Plus and phosphatase cocktail 1 (1:100). Sample protein concentrations were estimated using a Pierce protein BCA kit (Thermo Fischer Scientific) 9. 10µg (ERK), 25µg (Akt) and 35µg (GSK-3β) protein were prepared according to Fermentas DualColor protein loading buffer pack instructions (1 µL DTT and 5 µL loading buffer per 20µL total volume including protein sample and ddH2O; incubate for 5 minutes at 95-100 °C; spin a few seconds in a microcentrifuge). SDS-PAGE and Western Blotting. First, a 12% resolving gel (20 mL) was prepared by mixing 6.6 mL ddH2O, 8mL of 30% bis-acrylamide (Sigma), 5 mL 1.5 mol/L Tris-Buffer pH 8.8, 200 µL 10% Rosc-Schlüter et al. Online Supplement CVR-2010-1473.R2 SDS-solution (Fluka) in ddH2O, 16µl TEMED (Fluka) and 200µl 10% APS (ammonium persulfate, Sigma). The solution was transferred with a 10 mL pipette (Sarstedt) between the arranged glassplates (spacer plate and short plate) in the casting frame on the casting stand and gels polymerized within 30 minutes. To obtain an even gel, a thin layer of 70% ethanol (300-500 µL) was added on top of the gel, which was removed before the 10 mL 5% stacking gel [5.8 mL ddH 2O, 1.7 mL 30% bisacrylamide, 2.5 mL 0.5 mol/L Tris, pH 6.5, 100µl 10% SDS in water, 8µL TEMED and 100 µL 10% APS] was poured. Then the 5-, 10- or 15-well comb was carefully placed (gel polymerized within 20 minutes) and shortly before loading carefully removed. The gel apparatus was set up (MiniPROTEAN® Tetra Electrophoresis System, Bio-Rad) and filled with pre-cooled running buffer [1x TRIS-glycine buffer with SDS; 10 x Tris-glycine buffer: 250 mmol/L Tris-base (Sigma), 1.92 mol/L glycine, 20% SDS ]. Protein samples and prestained Protein Ladder were loaded in the slots with loading tips (Costar) and run with constant Volts (90V) till the tracking dye reached the bottom of the gel. Next, fiber pads, blotting paper (Whatmann, extra thick, pure cellulose) and PVDF-membranes were soaked in 1 used transfer buffer, whereas PVDF-membranes were first activated for 10 second in methanol (Chromasolv®, Sigma) and washed for 5 minutes in ddH2O. After running of the SDSPAGE, the stacking gel was removed; a sandwich of fiber-pads, filter paper, poly-aycrylamide gel, PVDF-membrane, filter paper and fiber pads was placed in the color coded holder cassette. The transfer apparatus was assembled (electrophoresis blotting module, 2 gel holder cassettes and the BioIceTM cooling unit; pre-cooled transfer buffer; Bio-Rad), placed in a Styrofoam box surrounded with ice cubes and a magnetic bone was added inside the buffer tank. The tank transfer system was plugged to a BioRad Power Pac 3000 for 2 hours with constant Volts (100V). After transfer, membranes were reactivated in methanol and blocked in 5% milk/TBST (TBS with 0.1% Tween-20, Sigma) for GSK3β (Ser9) or 4% BSA/TBST for 30 to 60 minutes. Effectiveness of protein transfer was controlled for by staining the gel with Coomassie Blue R (Sigma). Membranes were incubated with the primary antibody overnight, while gently shaking at 4°C (phospho and total ERK 1:5000, phospho and total Akt, total GSK3β 1:2500, phospho-GSK-3β 1:1500). Membranes were washed 3 times with TBST for 15 minutes. HRP-conjugated secondary antibody (in 4% BSA/TBST) was added and left for 45 min with gentle shaking at room temperature. Finally, membranes were washed 3 with TBST for 15 min, enhanced chemiluminscence was prepared according to the manufacturer’s protocols (Perkin Elmer Biosciences; Oxidizing Reagent Plus and Enhanced Luminol Reagent Plus in ratio 1:1) to detect immunoreactive bands with a blue X-ray film (Fisher Thermo Scientific) and a Curix Developer. Membranes were stained with Ponceau S to control for protein transfer and loading. The bands were analyzed using Image J v1.37 (http://rsb.info.nih.gov/ij/) 10. Flow Cytometry. The method was adapted from those described previously 11, 12. 25 mmol/L CMH2DCF-DA in DMSO was diluted to a 12.5 µmol/L working solution in pre-warmed (37°C) prfDMEM with L-glutamine (4 mmol/L) and Hepes (25mmol/L). Cardiomyocytes were treated with apocynin and DPI as described above and medium was exchanged for DCF working solution (2 mL/P60) for 20 min in the dark at 37°C and 5 % CO2. Cells were washed once with 7% FCS prfDMEM and twice with ice-cold PBS prior to detachment with accutase. Cells were transferred to polystyrene FACS tubes (BD Falcon), filtered through a 40µm cell strainer cap and centrifuged at 700 rpm at room temperature for 7 minutes. Pellets were re-suspended in 500µl prf-DMEM and kept on ice. CD29 (10 µg/mL for NRVM, 15 µg/mL for NMVM), H2O2 (100 µmol/L) and PMA (1 µmol/L) were supplemented prior to measurement at defined time points. The oxidative burst was measured with FL-1, 488nm excitation and 530 nm emission on a DAKO flow cytometer and DAKO Summit v.4.3 software. siRNA transfection. NRVM were transfected with 2μg/106 cells ON-TARGETPlus using the Amaxa Neonatal Rat Cardiomyocytes Nucleofector kit (VPE-1002), according to the manufacturer’s instructions, and incubated for 36 hours. Quantitative real-time RT-PCR. Total RNA was extracted using TriReagent (Sigma). RNA (2 μg) was reverse transcribed by random priming using a High Capacity cDNA reverse transcription kit (Applied Biosystems). The cDNA (5 μl) was analysed by real-time PCR on an ABI PRISM 7500 (Applied Biosystems, Rotkreuz, Switzerland), using TaqMan Universal PCR Master Mix (Applied Biosystems). The sequence of primers, all designed to be intron spanning, were 18S rRNA: 5`- Rosc-Schlüter et al. Online Supplement CVR-2010-1473.R2 CCATTCGAACGTCTGCCCTAT-3` and 5`-GTCACCCGTGGTCACCATG-3`; and NOX2: 5`CTGGGATGAATCTCAGGCCA-3` and 5`-TTCTCCTCATCGTGGTGCAC-3. 18S rRNA was used as internal standard. Relative levels of target mRNA were calculated using the ΔΔCT method. Cell viability. Cell viability was measured by an MTT-based colorimetric assay using Cell Proliferation Kit I (Roche) according to the manufacturer’s instructions. Statistical analyses. All statistical analyses were performed in Graphpad Prism Version 4.03 and pvalues below 0.05 were considered to indicate significant differences. Significance was tested in repeated measures 1-way ANOVA with post-test Bonferroni (for sample sizes ≤ 4) or Newman Keuls (for > 4 samples to compare) correction. Rosc-Schlüter et al. Online Supplement CVR-2010-1473.R2 References 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. Lam E, Zwacka R, Seftor E, Nieva D, Davidson B, Engelhardt J et al. Effects of antioxidant enzyme overexpression on the invasive phenotype of hamster cheek pouch carcinoma cells. Free Radiac Biol Med 1999;27:572-579. Zwacka R, Zhou W, Zhang Y, Darby C, Dudus L, Halldorson J et al. Redox gene therapy for ischemic/reperfusion of the liver reduces AP1 and NFB activation. Nat Med 1998;4:698-704. Davidson B, Allen E, Kozarsky K, Wilson J, Roessler B. A model system for in vivo gene transfer into the central nervous system using an adenoviral vector. Nat Genetics 1993;3:219-223. Thaik C, Calderone A, Takahashi N, Colucci W. Interleukin-1modulates the growth and phenotype of neonatal rat cardiac myocytes. J Clin Invest 1995;96:1093-1099. Lim C, Zuppinger C, Guo X, Kuster G, Helmes M, Eppenberger M et al. Anthracyclines induce calpain-dependent titin proteolysis and necrosis in cardiomyocytes. J Biol Chem 2004;279:8290-8299. Springhorn J, Claycomb W. Preproenkephalin mRNA expression in developing rat heart and in cultured ventricular cardiac muscle cells. Biochem J 1989;258:73-78. Xiang F, Sakata Y, Cui L, Youngblood J, Nakagami H, Liao J et al. Transcription factor CHF1/Hey2 suppresses cardiac hypertrophy through an inhibitory interaction with GATA4. Am J Physiol Heart Circ Physiol 2006;290:1997-2006. Masters JR, Stacey GN. Changing medium and passaging cell lines. Nat Protoc 2007;2:22762284. Kurien B, Scofield R. Protein blotting: a review. J Immunol Methods 2003;274:1-15. Burger W, Burge M. Digital imaging processing: An algorithmic introduction using Java: Springer-Verlag New York, 2007. Bass D, Parce J, Dechatelet L, Szejda P, Seeds M, Thomas M. Flow cytometric studies of oxidative product formation by neutrophils: a graded response to membrane stimulation. J Immunol 1983;130:1910-1917. Vowells S, Sekhsaria S, Malech H, Shalit M, Fleisher T. Flow cytometric analysis of the granulocyte respiratory burst: a comparison study of fluorescent probes. J Immunol Methods 1995;178:89-97. Rosc-Schlüter et al. Online Supplement CVR-2010-1473.R2 SUPPLEMENTAL FIGURES Supplemental Figure 1: CD29 induces time-dependent phosphorylation of cell-survival kinases. (A) ERK1/2, (B) Akt and (C) GSK-3β as detected by phospho-specific antibodies. NRVM were treated with 5 µg/mL activating α-CD29 antibody, unspecific IgG (IgG) or inhibitory anti-CD29 (iAb) antibody. Control (co) indicates baseline phosphorylation of the respective kinase. Kinase phosphorylation of the IgG and iAb treatment was not different from control in any case. Unless indicated otherwise, comparisons were not significant. Data are presented as mean ± s.e.m. * p < 0.05 and ** p < 0.01 (n=3 each). Rosc-Schlüter et al. Online Supplement CVR-2010-1473.R2 Supplemental Figure 2: GSK-3β is a downstream target of both, MEK/ERK and PI3K/Akt upon CD29 activation. Cardiomyocytes were treated with kinase inhibitors for 45 min prior to α-CD29 administration (5 µg/mL). The PI3K-inhibitors Wortmannin (W) and LY29400 (LY), the MEK1/2 inhibitor U0126 (U) and the Akt inhibitor Akti1/2 (Akti) were used. Phosphorylation-specific antibodies detected (A-D) ERK1/2 (Thr202/Tyr204), (E-H) Akt (Ser473) and (I-L) GSK-3β (Ser9), respectively. Data are mean ± s.e.m. Unless indicated otherwise, comparisons were not significant. * and # p < 0.05; ** and ## p < 0.01; *** and ### p < 0.001. (n=3 for I-L, H; n=4 for E-G, C; n=5 for A, B, D). Rosc-Schlüter et al. Online Supplement CVR-2010-1473.R2 Supplemental Figure 3: CD29-induced pro-survival signaling is ROS-dependent. NRVM were treated with MnTMPyP (MnT, 5 µmol/L, 45 min) prior to α-CD29 (5 µg/mL, 15 min). Aliquots of total cell lysate were analyzed with the respective rabbit α-phospho-antibodies [ERK1/2 (Thr202/Tyr204), Akt (Ser473) and GSK-3β (Ser9)]. α-CD29-induced phospho-ERK1/2 (A, n=10), phospho-Akt (B, n=6) and phospho-GSK-3β (C, n=4) signals were blocked by the SOD/catalase mimetic MnT. Data are mean ± s.e.m. Unless indicated otherwise, comparisons were not significant. *** p < 0.001, * p < 0.05, ## p < 0.01, # p < 0.05. Rosc-Schlüter et al. Online Supplement CVR-2010-1473.R2 Supplemental Figure 4: Adenoviral single-enzyme overexpression of SOD or catalase. (A-C) Cardiomyocytes were transfected with adenoviral constructs to overexpress SOD (Ad.Cu/ZnSOD1; SOD50), cat (Ad.Catalase, cat50) or β-galactosidase (Ad5CMVcytoLacZ; LacZ50) at 50 MOI 36 hours prior to α-CD29 administration (10 µg/mL, 15 min). Single enzyme overexpression of SOD significantly inhibited α-CD29 induced phospho-MEK1/2(A) and -GSK-3β (C), whereas Akt phosphorylation (B) was significantly increased. In contrast, cat efficiently inhibited phosphorylation of all three kinases (A-C). Phospho-specific antibodies were rabbit α-MEK1/2 (Ser217/221), α-Akt (Ser473) and α-GSK-3β (Ser9). Data are presented as mean ± s.e.m. Unless indicated otherwise, comparisons were not significant. * and # p < 0.05, ** and ## p < 0.01, *** p < 0.001 (n=5 for A and B, n=3 for C). Rosc-Schlüter et al. Online Supplement CVR-2010-1473.R2 Supplemental Figure 5: Apocynin and DPI impair CD29-induced survival signaling. Apocynin (500 µmol/L) was administrated overnight and DPI (10 µmol/L) for 45 minutes prior to α-CD29 (5 µg/mL, 15 min). Phospho-specific antibodies used were rabbit α-ERK1/2 (Thr202/Tyr204), α-Akt (Ser473) and α-GSK-3β (Ser9). Data are presented as mean ± s.e.m. Unless indicated otherwise, comparisons were not significant. ** p < 0.01, *** and ### p <0.001 (n=4 for C and F; n=5 for B; n=6 for A, D and E). Rosc-Schlüter et al. Online Supplement CVR-2010-1473.R2 Supplemental Figure 6: CD29-stimulation diminishes hydrogen peroxide-induced cell death in NOX2-competent cardiomyocytes. (A) NRVM were treated with hydrogen peroxide (H2O2; 500μM, 2.5 hrs) and viability was measured using an MTT-based colorimetric assay. Data are given as H2O2induced cell death (i.e. 100-viability in % vs. baseline). Pretreatment of NRVM with α-CD29 antibody (10 μg/ml, 15 min) markedly decreased H2O2-induced cell death (n=2 in triplicate). (B) NRVM were transfected with siRNA targeting NOX2 (siNOX2) or non-targeting siRNA (siCtr) as described and efficiency of NOX2 knock-down was assessed by quantitative RT-PCR (left panel, ***p<0.001, n=3). NRVM were exposed to H2O2 as per Figure S6A. Pretreatment with α-CD29 antibody (10-25 μg/ml, 15 min) diminished the H2O2–induced cell death in siCtr, but not in siNOX2 NRVM (right panel, n=3, *p<0.05).