Initiation of Protein Synthesis (4.1)
advertisement

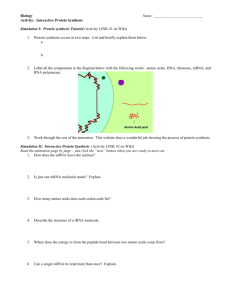
Initiation of Protein Synthesis (4.1) Lecture 4 Polypeptide Synthesis Overview Polypeptide synthesis proceeds sequentially from N Terminus to C terminus. Amino acids are not pre-positioned on a template. Proved by the classic experiment of Dintzis & coworkers in 1961. They synthesized hemoglobin in a test tube using a reticulocyte (red blood cell) system. They initiated protein synthesis and then added H3labeled leucine. They isolated hemoglobin, partially labeled with incorporated H3-leucine, and created peptide fragments (labeled a, b, c, d, e, f, g below) by digestion of hemoglobin with trypsin. Then they analyzed peptide fragments to determine the relative amount of radioactivity. The following results were obtained: http://bass.bio.uci.edu/~hudel/bs99a/lecture23/index.html (1 of 5)5/25/2007 9:36:32 AM Initiation of Protein Synthesis (4.1) http://bass.bio.uci.edu/~hudel/bs99a/lecture23/index.html (2 of 5)5/25/2007 9:36:32 AM Initiation of Protein Synthesis (4.1) Figure 4.1.1: Incorporation of radioactive leucine into hemoglobin after different periods of incubation. There is a gradient from the C-terminus to the N-terminus of the protein, which implies that synthesis takes place sequentially from N-terminus to C-terminus. As the time of incubation is increased, the total 3 amount of radioactivity is increased but the gradient remains. Results are expressed as the ratio of an H 14 label (tritium) in leucine to an internal control of C . Data of Dintzis (1961). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/index.html (3 of 5)5/25/2007 9:36:32 AM Initiation of Protein Synthesis (4.1) Ribosomes read mRNA in the 5'->3' direction, not the 3'->5' direction. To prove, one must know the Genetic Code. For example, AAA codes for Lys, AAC codes for Asn and CAA codes for Gln. In a cell-free protein synthesizing system, initiate protein synthesis by adding artificial mRNA, consisting of polyA with one C (cytosine) at the 3' end. Analyze the N and C termini of the peptide fragments. Nucleotide Sequence Protein Sequence Direction of Synthesis 5'-AAAAAA(AAA)nAAC3' Lys-Lys-(Lys)n-Asn 5' -> 3' 3'-CAAAAA(AAA)nAAA5' Gln-Lys-(Lys)n-Lys 3' -> 5' Next: Polypeptide Synthesis Overview (continued) http://bass.bio.uci.edu/~hudel/bs99a/lecture23/index.html (4 of 5)5/25/2007 9:36:32 AM Initiation of Protein Synthesis (4.1) Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/index.html (5 of 5)5/25/2007 9:36:32 AM Initiation of Protein Synthesis (4.2) Polypeptide Synthesis Overview (cont.) Active translation occurs on polyribosomes Electron micrographs of ribosomes actively engaged in protein synthesis revealed by "beads on a string" appearance. This implied that each mRNA transcript is read simultaneously by more than one ribosome. In fact, a second, third, fourth, etc. ribosome starts to read the mRNA transcript before the first ribosome has completed the synthesis of one polypeptide chain. Multiple ribosomes on a single mRNA transcript are called polyribosomes or polysomes. The individual ribosomes are separated by gaps of 50 Å to 150 Å. Thus, there is approximately one ribosome per 80 nucleotides. http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_2.html (1 of 4)5/25/2007 9:36:34 AM Initiation of Protein Synthesis (4.2) Chain elongation links the growing polypeptide to incoming aminoacyl-tRNA The newly formed polypeptide chain is attached to a tRNA molecule, not to a protein or mRNA molecule. To prove, use a cell-free protein synthesizing system with artificial mRNA, such as polyA. Add H3-Lys. Wash active ribosomes in high salt to dissociate and stop protein synthesis. Analyze results and find highest concentration of H3-Lys -- always with tRNA. From many such experiments, there are now known to be three tRNA binding sites on the ribosomes: A Site The ribosomal site which binds the incoming aminoacyl-tRNA (Acceptor site). P Site The site which holds the peptidyl-tRNA, that is the tRNA which is covalently linked to the growing polypeptide chain (Peptidyl site). E Site A site which transiently binds to the outgoing, deacylated tRNA (Exit site). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_2.html (2 of 4)5/25/2007 9:36:34 AM Initiation of Protein Synthesis (4.2) Three steps are involved in the synthesis of one polypeptide chain 1. INITIATION ❍ ❍ Assembly of active ribosome and the reading of the first mRNA codon (START codon). Occurs once per polypeptide chain. 2. ELONGATION ❍ Involves three distinct steps for each mRNA codon or amino acid in the new http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_2.html (3 of 4)5/25/2007 9:36:34 AM Initiation of Protein Synthesis (4.2) polypeptide chain: 1. Transfer of aminoacyl-tRNA from cytoplasm to A-site of ribosome. 2. Covalent linkage of new amino acid to growing polypeptide chain peptidyl transfer. 3. Movement of tRNA from A-site to P-site and simultaneous movement of mRNA by 3 nucleotides - translocation. ❍ Occurs multiple times per polypeptide. 3. TERMINATION ❍ ❍ Reading of final mRNA codon (STOP codon) and dissociation of polypeptide from ribosome. Occurs once per polypeptide chain. Next: Initiation of Protein Synthesis Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_2.html (4 of 4)5/25/2007 9:36:34 AM Initiation of Protein Synthesis (4.3) Initiation of Protein Synthesis INITIATION - Assembly of active ribosome and the reading of the first mRNA codon. Occurs once per polypeptide chain. The signals for initiation: mRNA Start Codon Prokaryotes: Eukaryotes: AUG (very rarely also GUG & UUG) AUG After protein is made, the N-terminal methionine is frequently cleaved off. Shine-Dalgarno Sequence To distinguish the start AUG codon from AUG codons which code for internal methionines or from a fortuitous AUG combination in another reading frame, the start AUG codon in prokaryotes is preceded by a highly conserved sequence at the 5´ end of the mRNA transcript which serves as a ribosome-binding site. The Shine-Dalgarno sequence is a purine-rich tract of 3-10 nucleotides and precedes the start AUG codon by approximately 10 nucleotides upstream on the 5´ side. The Shine-Dalgarno sequence forms base pairing with a highly conserved, pyrimidine-rich region of the 16S rRNA http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_3.html (1 of 4)5/25/2007 9:36:35 AM Initiation of Protein Synthesis (4.3) which is part of the 30S ribosomal subunit. This base-pairing properly aligns the start AUG codon in the P site of the 30S ribosomal subunit. In eukaryotes, protein synthesis usually begins with the first AUG codon from the 5´ end. Eukaryotic mRNA has a special cap at the 5´ end of the mRNA which is recognized by a cap-binding protein. Special Initiator tRNA Only one special tRNA is used to place the first amino acid, which is always a formyl-methionine (fMet): http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_3.html (2 of 4)5/25/2007 9:36:35 AM Initiation of Protein Synthesis (4.3) The initiator tRNA is called fMet-tRNA. It differs from all other tRNAs by one less base pair in the acceptor stem region and by three consecutive G:C base pairs in the anticodon region. These changes alter the 3-dimensional structure of the fMet-tRNA in subtle ways. fMet-tRNAfMet and Met-tRNAMet both recognize the AUG codon. A methionine is attached to both tRNAs by the same methionyl tRNA synthetase. However, once the methionine is attached to either tRNA, a special enzyme called formyl transferase, which recognizes only the initiator tRNAfMet adds a formyl group to the amide nitrogen. Because the amide N is now blocked from further reaction, this amino acid can only be positioned at the start of a polypeptide chain. Soluble Protein Factors In prokaryotes, there are three initiation factors: IF1, IF2, IF3, named in order of discovery, not in order of their function on the ribosome. The initiation factors are proteins that facilitate the assembly of an active http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_3.html (3 of 4)5/25/2007 9:36:35 AM Initiation of Protein Synthesis (4.3) ribosomal particle and the placement of the first fMet-tRNAfMet. In eukaryotes, there are 10 initiation factors, called eIF1, eIF2, etc. IF3 binds to 30S subunit and promotes the dissociation of the 30S subunit from the 50S subunit and the subsequent binding of the mRNA. IF1 helps IF3 by increasing the dissociation rate between the 30S and 50S subunit. IF2, in complex with GTP, binds to fMet-tRNAfMet and places it in the P-site of the 30S subunit. Then IF3 is released, causing GTP to hydrolyze to GDP, which in turn releases IF2-GDP. GTP hydrolysis promotes the release of IF1 and the subsequent association of the 30S subunit with the 50S subunit. Next: Schematic of Initiation Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_3.html (4 of 4)5/25/2007 9:36:35 AM Initiation of Protein Synthesis in Prokaryotes (4.4) Formation of the 70S initiation complex Schematic of the initiation of protein synthesis in prokaryotes http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_4.html (1 of 3)5/25/2007 9:36:36 AM Initiation of Protein Synthesis in Prokaryotes (4.4) 1. Binding on IF1 & IF3 to an empty 70S ribosome dissociates it into 50S and 30S subunits. 2. a. 5' region of mRNA binds to free 30S ribsomal subunit, IF3 is released. b. Initiator fMet-tRNAfMet carried by IF2-GTP binds to P site of 30S ribsomal subunit. 3. Free 50S ribosomal subunit binds, IF1 & IF2-GDP + Pi are released. Certain details of the process remain uncertain. For example, the exact order of binding of IF1, IF2, IF3, tRNA, and mRNA is unclear. The above is one current model (from the text book). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_4.html (2 of 3)5/25/2007 9:36:36 AM Initiation of Protein Synthesis in Prokaryotes (4.4) Next: Initiation of translation in eukaryotes Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_4.html (3 of 3)5/25/2007 9:36:36 AM Initiation of Protein Synthesis in Eukaryotes (4.5) Schematic of the initiation of protein synthesis in eukaryotes http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_5.html (1 of 2)5/25/2007 9:36:37 AM Initiation of Protein Synthesis in Eukaryotes (4.5) Note that the process is in principle very similar to prokaryotic initiation. Major differences from prokaryotic initiation are associated with the m7G cap of the mRNA and its interaction with the cap-binding protein (CBPI) and eIF4F (CBPII) instead of Shine-Dalgarno sequencemediated alignment to properly position the first AUG codon in the P site of the ribosome. Next: Summary Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_5.html (2 of 2)5/25/2007 9:36:37 AM Summary (4.6) Summary ● ● ● ● ● Polypeptide synthesis proceeds sequentially from N terminus to C terminus Ribosomes read mRNA in the 5'->3' direction, not the 3'->5' direction Active translation typically occurs on polyribosomes Chain elongation links the growing polypeptide to incoming aminoacyl-tRNA Three steps are involved in the synthesis of one polypeptide chain of N residues: 1. INITIATION (1 x) 2. ELONGATION (N-1 x) 3. TERMINATION (1 x) ● Initiation in prokaryotes requires: 1. One empty 70S ribosome 2. Three initiation factors: IF1, IF2, IF3 3. mRNA with Shine-Dalgarno sequence and START codon (AUG) 4. Initiator fMet-tRNAfMet 5. One GTP ● Initiation in eukaryotes is similar, but more complicated Next lecture: Elongation & Termination of Protein Synthesis http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_6.html (1 of 2)5/25/2007 9:36:38 AM Summary (4.6) Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_6.html (2 of 2)5/25/2007 9:36:38 AM