Pglo Bacterial Transformation
advertisement

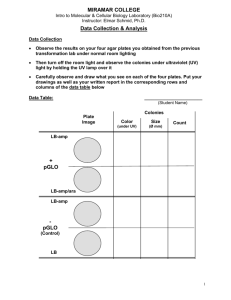
GENETIC TRANSFORMATION OF BACTERIA WITH THE GENE FOR GREEN FLUORESCENT PROTEIN (GFP) LAB BAC3 Adapted from "Biotechnology Explorer pGLO™ Bacterial Transformation Kit Instruction Manual". (Catalog No. 166-0003-EDU) BIO-RAD Laboratories, 2000 Alfred Nobel Drive, Hercules CA 94547. STANDARDS ADDRESSED 3.1.10A/3.1.12A Apply concepts of systems and control at the molecular level to assess the outcome of bacterial transformation. 3.2.10B/3.2.12B Describe the transformation results using precise quantitative and qualitative skills based on observations. Evaluate the experimental data correctly within experimental limits. 3.3.10C/3.3.12C Describe how genetic information is inherited and expressed at t the molecular level. KEY CONCEPTS: Genetic transformation, plasmid DNA, cloning, restriction enzymes, antibiotic selection, gene regulation, transcription, protein expression INTRODUCTION Genetic transformation is the process by which a gene or genes from one organism are transferred to another organism via an engineered molecule of DNA. If the procedure is successful, the organism is capable of producing the protein encoded by the transformed gene, thus creating a genetic change. Genetic transformation is commonly used in biotechnology. In agriculture, transformation is used to confer genes for pest, frost and spoilage resistance. Transformation of the human insulin gene into bacteria has allowed for production of the protein on a large scale. 1 To aid in bioremediation of oil spills, bacteria are transformed with genes that allow them to digest toxic components of the oil. 2 The procedure contained in this lab will allow for the transformation of bacteria with a gene from the bioluminescent jellyfish, Aequorea victoria. A successful transformation will result in the expression of the green fluorescent protein (GFP) in the bacteria, causing them to glow bright green under long-wave UV light. 1 2 http://www.littletree.com.au/dna.htm http://www.epa.gov/ORD/NRMRL/lrpcd/esm/oil_spill_bioremediation_researc.htm _______________________________________________________________________ Westminster College SIM MICRO3-1 Transformation of Bacteria with GFP Transformation and Antibiotic Selection: Genetic transformation in this laboratory will be facilitated by using the pGLO plasmid (see below). A plasmid is a circular, selfreplicating DNA molecule which can be contained in a bacterial host cell without interfering with the function of the bacterial chromosome. Bacteria are capable, on their own, of randomly acquiring small pieces of DNA from their environment, but the process is inefficient. The transformation protocol in this lab uses a chemical, calcium chloride (CaCl2), plus heat to increase the efficiency of DNA uptake by the bacterial cell. Even with the chemical transformation procedure, not every bacterial cell will incorporate the pGLO Figure 1 plasmid into the bacteria, not every cell will receive a copy of the plasmid. To isolate only the cells containing the pGLO DNA, the plasmid contains the beta-lactamase gene which encodes for an ampicillin resistance (Ampr) protein. After the transformation, the cells are grown on a solid medium called an agar plate. This medium will contain the antibiotic ampicillin. In the presence of the ampicillin, only the bacteria containing the pGLO plasmid will have the Ampr protein which will break down the antibiotic, and be able to grow (Fig. 2). This process is called antibiotic selection. Figure 2. Transformed Bacterium: This is a diagram of what is occurring inside a bacterium transformed with the pGLO plasmid. The bacterium is plated on agar medium containing ampicillin. To grow, the bacterium must contain a pGLO plasmid and be expressing the ampicillin resistance protein, β-lactamase (Ampr). The ampicillin is inactivated by the β-lactamase protein, allowing the bacterium to grow on medium containing the antibiotic. = pGLO plasmid =Ampr = transcription & translation Bacterial chromosome pore = ampicillin/ inactivation Cloning a Gene: Plasmids can be engineered to carry a variety of genes that are not endogenous to the host cell, like the GFP gene. A plasmid usually starts out as a very small piece of DNA that contains a replication origin, an antibiotic resistance gene and a cloning region, an area of the DNA that has multiple unique restriction enzyme sites. Westminster College SIM BAC3-2 Transformation of Bacteria with GFP Restriction enzymes are proteins which recognize specific DNA sequences and will cleave, or cut, the DNA backbone at these sequences. Once the DNA backbone is cleaved, it is possible to add, or clone, new DNA into this site. The diagram of the pGLO plasmid (Fig. 1) shows restriction sites for the enzymes NdeI, EcoRI, and HindIII, enzymes that are part of the original cloning site on the plasmid. The pGLO plasmid also carries the gene araC, which produces a protein needed for transcription of genes in the presence of arabinose sugar. The purpose of this protein is discussed below. Transcriptional Gene Regulation: In many cases, a researcher may want to control when a cloned gene is producing mRNA, and the corresponding protein. Proteins called transcription factors are frequently used by cells to turn transcription “on” or “off” depending on environmental conditions. The transcription, or production of mRNA, of the pGLO gene is controlled by using a promoter that is only active in the presence of the sugar arabinose (Fig. 3). The AraC protein, encoded by the araC gene on the pGLO plasmid, is the transcription factor necessary for this control. This protein is bound at the pGLO promoter site, but without arabinose is in the incorrect conformation, or shape, to recruit RNA polymerase and initiate transcription. (See figure below) In the presence of arabinose, the sugar binds to the AraC protein and changes its conformation so that in combination with RNA polymerase, transcription is initiated and an mRNA transcript is produced. In bacteria, transcription and translation, or protein synthesis, occurs simultaneously, and the GFP protein is produced. Westminster College SIM BAC3-3 Transformation of Bacteria with GFP Fig. 3 Regulation of the GFP Gene promoter AraC GFP A. In a cell with no arabinose present, the AraC protein, bound to the DNA, is in the wrong configuration to recruit RNA polymerase to the transcriptional promoter. This means the promoter is “off” and there is no transcription of the GFP gene. arabinose GFP AraC B. A bacterium is able to take up arabinose when it is present in the environment. Once inside the cell, arabinose binds to the AraC protein, changing its shape, or conformation, on the DNA. This new protein conformation recruits RNA polymerase to the transcriptional promoter. RNA Pol AraC GFP C. The binding of the RNA polymerase turns the promoter “on” and allows transcription of the GFP gene. mRNA transcript RNA Pol AraC GFP D. Transcription of the GFP produces an mRNA transcript. In bacteria, transcription and translation (synthesis of proteins) can occur simultaneously, and the GFP protein is produced within the cell. In this lab, it is important to confirm which cells have received the plasmid, and under which conditions the β-lactamase and GFP proteins are being produced (Fig. 4). When the pGLO transformations are plated on agar medium containing ampicillin and arabinose, a series of controls will be plated as well (Fig. 5). Two transformations will be performed: one with pGLO plasmid (+pGLO) and one without the plasmid present (pGLO). A portion of the –pGLO transformation is plated on an agar medium without ampicillin or arabinose. This control is to be sure the bacteria are viable after the chemical and heat transformation procedure. This plate should be covered with a bacterial “lawn”. Another portion of the –pGLO transformation is plated on an agar plate Westminster College SIM BAC3-4 Transformation of Bacteria with GFP Figure 4. Expression of GFP: This is a diagram of what is occurring inside a bacterium transformed with the pGLO plasmid. The bacterium is plated on agar medium containing ampicillin and arabinose. To grow, the bacterium must contain a pGLO plasmid and be expressing the ampicillin resistance protein, β-lactamase. In the presence of arabinose, transcription of the GFP gene is enabled, resulting in expression of the GFP protein. = pGLO plasmid =Ampr = GFP protein Bacterial chromosome = transcription & translation pore containing ampicillin. No bacteria should grow on this plate. If it does, it means the bacterial culture has acquired resistance and is no longer suitable for use in this experiement. The +pGLO transformation is also plated on two different types of media. The first portion of the transformation is plated on agar containing ampicillin only. This control proves that the transcriptional control of the GFP gene is intact, and no GFP protein is produced in the absence of the arabinose sugar. The final plate is the experimental plate, containing both ampicillin and arabinose. The bacteria on this plate are the only ones that should glow when exposed to long-wave UV light. Figure 5: Controls for transformation of the pGLO plasmid LB = positive control for cell growth LB amp= negative control for cell growth LB amp = transformation; Ampr w/o GFP expression LB amp/ara = transformation; Ampr and GFP production Westminster College SIM -pGLO Plasmid HB101 cells +pGLO Plasmid HB101 cells BAC3-5 Transformation of Bacteria with GFP PROCEDURES It is important to remember that sterile technique is extremely important for this lab. All the materials provided are sterile and proper handling of these supplies and reagents should result in minimal contamination problems. Lab Station Materials 2 sterile 1.5 mL microtubes Sterile transfer pipets (10) Sterile loops (8) Ice water baths LB plate (black stripe) LB amp/ara plate (green stripe) Sterile LB broth (600 µL) Lab tape Sharpie pen Sterile CaCl2 transformation solution (600 µL) pGLO plasmid DNA Foam microtube rack 2 LB amp plates (red stripe) Water bath @ 42°C 30°- 37°C Incubator (optional) Microtube rack Procedure 1. There are 2 sterile 1.5 mL microfuge tubes at the lab station. Label one tube “+pGLO”, the other “-pGLO”. 2. Locate the tube labeled “Transformation Solution”; this contains a solution of CaCl2. With a sterile pipet, aliquot 250 µL transformation solution to both the -pGLO and +pGLO tubes. Place both tubes in an ice water bath. 3. The lab station will contain a starter plate of HB101 bacteria. These bacteria contain only chromosomal DNA. Take notes on how these bacteria look in visible and UV light for comparison with the transformed bacteria later in the lab. The lab station will also include pre-packaged sterile loops (look like a very small soap bubble wand). Remove a sterile loop from the package being careful not to touch the loop on anything outside the bag. Remove the cover from the starter plate and pick a single colony of the HB101 bacteria. Immediately place the loop into the transformation solution in the tube marked “+pGLO” and spin the loop until the entire colony is dispersed into the liquid. Check to be sure there are no bacterial clumps floating in the transformation solution; cell clumps will negatively affect the efficiency of the transformation. Place the tube back in the ice water bath when the cell suspension is complete. 4. Using a new sterile loop, repeat the procedure in Step 3 for the “-pGLO” tube of transformation solution. 5. Inspect the pGLO plasmid solution with the UV lamp provided and note your observations. What do you expect to see at this step? Take the 2-20 µL pipettor, set it to Westminster College SIM BAC3-6 Transformation of Bacteria with GFP 5 µL. Carefully remove 5 µL of the pGLO plasmid solution and add the DNA into the “+pGLO” cell suspension. Mix the plasmid with the bacterial cells by tapping the tube gently on the bench top. Close the tube and return it to the ice water bath. Why do you not add plasmid DNA to the tube labeled “-pGLO”? 6. Incubate both the “+pGLO” and the “-pGLO” tubes in the ice water bath for 10 min. Make sure the bottom of the tube is pushed through the foam rack and is in contact with the ice water. 7. While the tubes are incubating on ice, label the 4 agar plates at your lab station as follows: Color code: red stripe green stripe red stripe black stripe Be sure to label the bottom of the plates (the portion containing the agar). It is very important to label each plate correctly! Remember, the color code on the side of the agar plates indicates which medium already contains the ampicillin for antibiotic selection and arabinose for GFP expression. It will be confusing to analyze the results if you have mislabeled the plates! 8. To transform the plasmid DNA into the bacteria, the cells must undergo a heat shock. This is performed by removing the foam rack from the ice water bath and placing it rapidly in a 42°C water bath. Incubate the tubes at 42°C for exactly 50 seconds. Again, it is important that the tubes are pushed down in the rack so that the bottom of the tubes have optimal contact with the 42°C water. After the heat shock, immediately place the foam rack back in the ice water and incubate for a further 2 min. 9. Remove the foam rack from the ice water bath and place the tubes in the microtube rack on the bench. Add 250 µL of LB broth to each tube, close the cap and gently tap the “+pGLO” and “-pGLO” tube to mix the contents. Incubate the tubes for 10 min at room temperature. This step of the procedure allows the cells to recover from the heat shock treatment before performing the next part of the experiment. It also lets the cells that have acquired a pGLO plasmid begin to express the β-lactamase protein (for ampicillin resistance) before the cells are placed on plates that contain ampicillin. Westminster College SIM BAC3-7 Transformation of Bacteria with GFP 10. Using a new sterile pipet for each tube, pipet 100 µL of the transformation and control suspensions onto the appropriate plates 11. Spread the suspensions evenly around the agar plate by quickly sliding the flat loop surface back and forth across the plate surface. Turning the plate in a circular motion with your fingers while swishing the loop back and forth aids in spreading the bacterial suspensions evenly. Do not press down too firmly or you will gouge the surface of the agar plate. This can complicate both the growth and analysis of the bacteria on the plates. Remember to use a new sterile loop for each plate! Let the plates sit on the bench for 2-3 minutes to allow the suspensions to soak into the plate. 12. Stack the plates and tape them together. Label the tape with the group name and class period, if necessary. Turn the plates upside down (with agar at top of plate) and place in a 37°C incubator overnight. If an incubator is not available, the plates may be grown on the bench top for 2-3 days. Stack of upsidedown plates: Incubate at 37°C Westminster College SIM BAC3-8 Transformation of Bacteria with GFP DATA & ANALYSIS SHEETS Name:________________________ Class: ________________________ Date: ________________________ Day 1: Predictions & Questions a. Which agar plate should contain the most colonies? The least? What do these two control plates tell us? b. On which plate(s) should the bacterial colonies glow when examined with the UV light? c. One of these plates will contain bacteria that contain the pGLO plasmid, but will not fluoresce when examined with the UV light. Which plate is this, and why won’t the colonies glow? d. In the “-pGLO” tube, there was no DNA present to transform into the cells. Why does this tube need to go through the heat shock procedure? Day 2: Data Collection and Analysis of Results Observe the results you obtained from the transformation lab under normal room lighting. Then turn out the lights and hold the ultraviolet light over the plates. 1. Carefully observe and draw what you see on each of the four plates in the table below. Record your data to allow you to compare observations of the “+ pGLO” cells with your observations for the non-transformed E. coli. Write down the following observations for each plate. 2. How much bacterial growth do you see on each plate, relatively speaking? 3. What color are the bacteria? 4. How many bacterial colonies are on each plate (count the spots you see)? Westminster College SIM BAC3-9 Transformation of Bacteria with GFP Data Collection Table: General growth # bacteria color (vis/UV light) Analysis of Data: 1. On Day 1, you made predictions about what would happen on each plate in terms of bacterial growth. How does the data collected compare with your predictions? If there are significant differences, what do you think caused them? Westminster College SIM BAC3-10 Transformation of Bacteria with GFP 2. Both of the plates that contain the “-pGLO” samples are controls. What information is given by these plates that aids in interpreting the “+pGLO” plates? a. “-pGLO” on LB agar: b. “-pGLO” on LB amp agar: 3. Very often an organism’s traits are caused by a combination of its genes and its environment. Think about the green color you saw in the genetically transformed bacteria: a. What two factors must be present in the bacteria’s environment for you to see the green color? (Hint: One factor is in the plate, the other is in how you observe the bacteria.) b. What do you think each of the two environmental factors you listed above are doing to cause the genetically transformed bacteria to turn green? c. What advantage would there be for an organism to be able to turn on or off particular genes in response to certain conditions? Westminster College SIM BAC3-11 Transformation of Bacteria with GFP 4. Your and your partner performed the pGLO transformation lab, correctly plated the bacteria after transformation and placed your carefully labeled plates in the 37°C incubator. Unfortunately, as an April Fools Day prank, another student wiped all the identifying labels off the plates with an organic chemical. Assume the transformation was successful. Based on your predictions and the data you collect from the plates, is there a way to deduce which plate is which? Which plates contain bacteria with the transformed pGLO plasmid? Which plate is the LB agar? Which plates contain ampicillin, arabinose or both? What factors help you make your decision? # bacteria 88 Appearance of Bacteria room light under UV tan green “Lawn” tan white No growth -- -- tan white 93 Westminster College SIM pGLO? Amp/Ara? BAC3-12 Transformation of Bacteria with GFP EXTENSION ACTIVITY Calculate Transformation Efficiency Transformation efficiency is a way of calculating how many bacterial cells actually received the pGLO plasmid during the transformation procedure. In many experiments, like gene therapy, optimizing the number of cells receiving new genetic material is very important. There are two important pieces of information you will need to determine the transformation efficiency. 1. The total number of green fluorescent colonies growing on the LB amp/ara plate. This is data you have already obtained when analyzing your plates. _____ = # of green fluorescent colonies 2. The total amount of pGLO plasmid DNA in the bacterial cells spread on the LB amp/ara plate. This part of the calculation has a several steps which will determine the amount of pGLO plasmid spread on the LB amp/ara plate. a. The stock solution of pGLO plasmid DNA is: 80 ng μL or 0.08 μg μL b. The total amount of pGLO plasmid added to the transformation tube is calculated by multiplying: (concentration of DNA in µg/µL) x (volume of DNA in µL) = DNA in µg Or 5 µL X 0.08 μg = ______µg = Total pGLO in transformation μL c. Since not all the DNA you added to the bacterial cells will be transferred to the agar plate, you need to find out what fraction of the DNA was actually spread onto the LB amp/ara plate. To do this, divide the volume of the transformation solution spread on the LB amp/ara plate by the total volume of liquid in the test tube containing the DNA. Calculate the total transformation solution volume: CaCl2 Solution = pGLO plasmid solution = LB broth = Westminster College SIM µL Volume on plate = ____ µL µL µL µL total volume BAC3-13 Transformation of Bacteria with GFP Fraction of transformation = Volume spread on LB amp/ara plate (µL) = ______ used Total transformation volume in tube (µL) d. The fraction of transformation solution gives a percentage. To calculate the amount of pGLO plasmid on the plate, multiply this percentage (from c.) by the total µg pGLO DNA (from b.). (Fraction of transformation) X (total µg pGLO) = pGLO plasmid on plate (µg) ____________ X __________µg = _____ µg e. The final calculation is to determine the number of colony forming units (cfu) per ug DNA transformed. Divide the number of colonies counted on the LB amp/ara plate (see Step 1) by the µg DNA determined in 2d. (above). # colonies = _____________ = __________ cfu µg DNA µg µg The transformation efficiency will vary greatly depending on the method of transformation used. It is almost always expressed in scientific notation. For example, if you calculate a transformation efficiency of 550 cfu/µg, then you would write this as: 5.5 x 102 cfu/µg Westminster College SIM BAC3-14 Transformation of Bacteria with GFP Flow Chart of pGLO Transformation Protocol = Overnight step Label two tubes “+pGLO” and “-pGLO”. Add 250 µL transformation solution to each. Place tube in ice water bath _ + _ + Inoculate each tube with HB101 bacterial colony from starter plate. Disperse bacteria evenly and place tubes back in ice water bath _ + Add 5 µL of the pGLO plasmid to the “+pGLO” tube. Mix well and place tube back in ice water bath. -pGLO LB Incubate tubes on ice for 10 min. -pGLO LB amp Label LB agar plates during incubation. +pGLO LB amp +pGLO LB amp/ara + _ Incubate tubes on ice for 2 more min. Heat shock transformation mix tubes in 42°C water bath for 50 sec. + _ Remove tubes from ice & place on bench. Add 250 µL LB broth to each. Incubate for 10 min at room temperature. (Go to next page.) Westminster College SIM BAC3-15 Transformation of Bacteria with GFP Use a sterile spreader to plate 100 µL of appropriate transformation mix on each labeled agar plate. _ LB = positive control for growth LB amp= negative control for growth + LB amp = transformation; Ampr w/o GFP expression LB amp/ara = transformation; Ampr and GFP production Grow plates overnight at 37°C. Do not overgrow or individual colonies may be difficult to count. LB = positive control for growth LB amp= negative control for growth LB amp = transformation; Ampr w/o GFP expression LB amp/ara = transformation; Ampr and GFP production Analyze results of plates in the following class period. Westminster College SIM BAC3-16