Lab 3 Handout - York College of Pennsylvania
advertisement

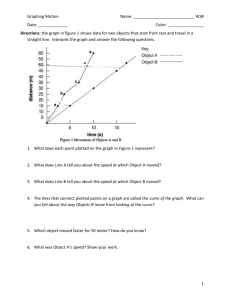
BIO 300 Ecology 1 LAB 3 SPECIES AREA CURVES AND HABITAT DIVERSITY Introduction: Scientists frequently want to be able to characterize a population or to compare populations. For example, they may want to know the density (number per unit area) of beech trees in a forest. Epidemiologists may want to know whether high blood pressure is more frequent in persons living close to metropolitan airports than in persons living away from them. In some circumstances it may be possible to deal with complete populations. You could census all of the students at York College or count all of the trees in a small tract of woods. For example, we might visit a small island and live-trap and weigh every mouse on it. We would then know, among other things, the total population size and weight of that population of mice. Traits of whole populations are called parameters. Frequently, we cannot measure every individual in a population due to such considerations as time and money. Instead, we sample a fraction of the population. We then generalize from the traits the sample possesses (called statistics) to the parameters of the whole population. If the sample is representative of the whole population, we may do this and draw conclusions that (after allowing for some statistical uncertainty) will be correct. Two features determine whether a sample will be representative: 1) the sample must be unbiased, and 2) it must be adequate in size. During the first lab exercise, we touched upon the issue of unbiased sampling by randomly sub-sampling an area. Today, we will address the adequacy of sample size and species diversity. Today’s Objectives: 1) Use three methods to judge the adequacy of sampling (i.e. sample size) of Indian tobacco (Lobelia inflata). 2) To examine the arthropod communities inhabiting two different habitats. 3) To develop species diversity indices for the habitats sampled. 4) To measure the degree of similarity of the two habitats. 5) To related structural complexity of the habitats with arthropod species diversity. Adequacy of Sampling: Sometimes it is not possible to specify ahead of time how large a sample is needed to be adequate. In general, the larger the sample size, (the more quadrats or other sample unit used) the more likely it will be adequate. In general, a simple homogeneous area will require a fewer samples than a complex or heterogeneous one. The following are three ways to assess the adequacy of sampling populations. 1. The species:area curve. A plot of the accumulated number of species against the size of the sample (for example, the number of quadrats) is called a species:area curve. This is briefly addressed in Krohne (pp. 298-299). The specific aspect of adequacy to which it applies is whether our sample has been large enough to include most of the species in the community we are studying. The concept is that in our first quadrat we will have all new species. In the second quadrat we will probably repeat some species from the first quadrat, but we will observe several new ones as well. As we continue to sample, we will reach a point where our samples have included all the common species and many of the rarer ones, so that each new quadrat adds little to the accumulated species total. Our plot of cumulative number of species begins to level off (Figure 5-1 of this handout). There is no level at which species registration will be adequate for every purpose; a fairly widely used rule is that species sampling will be assumed to be adequate when a given percentage increase in sample size Rev. 9/2005 BIO 300 2 Ecology produces less than the same percentage increase in number of new species. A mechanical way of determining this is described below. Refer to figure 5-1 below. From Brewer and Mcann (1982) 1. Plot the cumulative number of species for each sequential quadrat on the y-axis against the number of quadrats sampled on the x- axis. If we have completed enough samples (at least for plants) we should observe a sharp rise and a leveling off characteristic of species:area curves . 2. Now draw a line with a slope of 1 that starts at zero on both axes (the origin). Along this line the addition of one new sample adds one new species. a. For any portion of the species:area curve that rises more steeply than this line, species are being added at a greater rate than sample area is being added. b. For any portion of the curve that has a slope flatter than the line, species are being added at a lower rate. 3. Draw a line parallel to this line and just touching the species:area curve. At this point (where the straight line is tangent to the curve) one new species is being added with each new sample. 2) The Performance Curve: A performance curve plots the mean value of some trait against sample N size. Each consecutive point on the Y-axis is the mean value of i samples, where Yi = ∑ ni , i =1 beginning with i = 1 and N= the total number of samples taken. This could be density, biomass, weight, or any of a variety of other measurements. The mean value of the first one or two samples are not likely to approximate the population mean, so that the early part of the performance curve tends to be jagged. As more samples are added, the curve flattens out as the mean value of your sample comes closer to the true population mean. When the fluctuations produced by a new sample become very small we assume that we are estimating a mean that is close to the true population mean. (See figure on next page.) . Rev. 9/2005 BIO 300 3 Ecology From Brewer and McCann (1982) 3) Two-step Sampling: Although interpretation of the performance curve could be made objective, a better way to obtain a more rigorous statistical approach is to employ two-step sampling. For example, we could sample ten quadrats, and then based on how variable those ten quadrats are, calculate how many more quadrats we will need to achieve some certain level of statistical reliability. The method depends on the statistical concept of the confidence interval. This is the interval within which we are confident, with some specified probability that a population trait lies. Here we are interested in specifying the confidence interval for the mean density of a population. The formula for calculating the confidence interval: the lower limit of the confidence interval is - x − K (S / N ) the upper limit of the confidence interval is - x + K (S / N ) where x is the sample mean, S the sample standard deviation, N the sample size (number of samples, or quadrats), and K the normal curve variate for a particular probability. For example, suppose we have a sample with the following traits: N = 200, x = 31.7, and S = 1.81. We wish to be 90% certain that the true mean of the population lies within limits we select. The K value for the 90% level is 1.645. This is looked up in a table; however, the only probabilities that we are likely to want, and the corresponding K values are, 90% - 1.645 95% - 1.960 99% - 2.576 Using a K value of 1.645, we have confidence limits of 31.7 – 1.645(1.81/ 200 ) = 31.49, and 31.7 + 1.645(1.81/ 200 ) = 31.91 Based upon our sampling, we say that we are 90% confident that the true population mean lies within this range. In other words, when we make the statement that the true population mean lies in this interval, we will expect to be right 90 times out of 100. Rev. 9/2005 BIO 300 Ecology 4 The important feature of the confidence interval for this method of determining adequacy of sampling is that any increase in sample size will decrease the size of the confidence interval. (This should be clear from the fact that we divide by sample size in the calculations.) We can use this relationship to assess the number of samples we need to attain a given level of precision. Suppose we have sampled 20 quadrats and obtained a mean of 8.0 and a standard deviation of 3.0. We want to be sure with 95% confidence that our sample mean is within ± 1.0 of the population mean. Are 20 samples enough, or do we need to continue sampling? If so, how many more will we need? We want a sample size that satisfies this relationship: 1.0 = 1.96 (3.00)/ √N) where 1.96 is the K value for the 95% confidence interval. The general form of the equation is: L = K (S/√N) Where L is the confidence interval and the other symbols are the same as above. So we solve for N S2 x K 2 N= L2 In this example, N= 3 2 x 1.96 2 9 x 3.84 = = 34.6 1 12 From this we conclude that we will need 35 samples to obtain a sample mean within ± 1 of the population mean. Our sample of 20 is not enough. We will need to sample 15 more quadrats. Additional methods for characterizing a community. Species Diversity Indices Species diversity is a characteristic unique to the community level of organization. The diversity of species within a given habitat is known as alpha diversity. Beta diversity, in contrast, describes the degree of change in species from one habitat to another. Alpha diversity combines two distinct aspects of the species composition of communities: number of species and equitability or evenness of abundance. These two aspects of diversity can be described separately, but they are often considered together as indices of heterogeneity. Krohne discusses measures of species diversity on pp. 296-298. Rev. 9/2005 BIO 300 5 Ecology One way to evaluate communities would be to compare the number of species within each community. This is known as species richness. Each species, however, is not likely to have the same number of individuals in that community. One species might be represented by 1000 individuals, and another by 200, and a third by a single individual. The distribution of individuals among species is called species evenness, or species equitability. Evenness is maximized when all species have the same number of individuals. For this lab, you will be concentrating on calculating species richness and species evenness. The following is information on combining both those measures which you are required to know. Species diversity is a combination of richness and evenness; it is species richness weighted by species evenness, and there are formulas that permit the diversity of a community to be expressed in a single number. Diversity indices are less frequently called indices of heterogeneity. Shannon-Weaver index – the most widely used index of heterogeneity. It is derived from a function used in the field of information theory to describe the average degree of uncertainty of occurrence of a particular symbol at a certain point in a message, and consequently, the amount of information conveyed by its presence. (The stuff you learn in spy school.) A secrete message such as dddddd has little uncertainty as to what the next letter will be, knowing what the previous letters were. In our case, we want to know what the next species will be. Where all the bits of information are the same, that is, where the species diversity is low, it is easy to predict the next bit. Where the bits of information are different, (e.g. askritnv), it is hard to predict the next bit, and diversity is high. As a diversity index for biotic communities, the Shannon-Weaver index describes the average degree of uncertainty of predicting the species of an individual picked at random from the community. This uncertainty increases both as the number of species increases and as the individuals are distributed more and more equitably among the species already present. An index of diversity provides a crude, single figure representation of the number of species and their evenness. The most widely used is the Shannon-Weaver index. S The general form of the index, H′, is H ′ = −∑ ( pi )(log pi ) Where pi = decimal fraction of i =1 individuals belonging to the ith species. (e.g. if the fraction, or proportion of individuals is 50%, then it is expressed as 0.50). The Shannon-Weaver index varies from a value of 0 for communities with only a single species to high values for communities having many species, each with a few individuals. It is important to emphasize that species richness and diversity are quite different. Although richness and diversity are often positively correlated, environmental gradients do exist along which a decrease in richness is accompanied by an increase in diversity. For example, Species A B C D E F G H’ Community A Community B # of individuals 10 2 1 2 1 2 1 2 1 2 1 1 0.579 .699 Community A has higher species richness, but community B has greater species evenness, hence it scores a higher diversity index. Rev. 9/2005 BIO 300 6 Ecology The deciduous forests of the eastern United States are of intermediate diversity, with H′ values for tree species ranging only from a little over 3.0 for the mixed mesophytic forest in the Cumberland and Allegheny Mountains to values less than 2.0 at the western and northern edge. Out west, a value of 7 could be expected in the more species rich forests of the Siskiyou Mountains of Oregon and California. For this lab, you will calculate an evenness index, which is very similar to the Shannon-Weaver index. The Shannon-Weaver index is the numerator of the evenness index on page 51 of Allen et al. (1975). "THERE ARE TWO WAYS TO CALCULATE THIS NUMERATOR (i.e. SHANNON-WEAVER INDEX)! (Note: These are not the equations for evenness! The calculations here are for the numerator of the evenness index. See page 2 in Allan et al. or page 7 of this handout for calculating evenness.) S 1) To calculate H’ you could use the following equation H ′ = −∑ ( pi )(log pi ) . This is the i =1 traditional format. It is easy to calculate if you are comfortable using spreadsheets. 2) If your mode of math is slide rule, calculator or paper and pencil, then this is the method for you! The above method involves too many calculations (such as converting the number of individuals to proportions) and there is always the chance for rounding errors. An algebraic manipulation of the traditional Shannon-Weaver equation (above) yields; H’ = [(N log N) - Σ[ni log ni]]/N Where N = to total number of plants in the sample And ni = the number of individuals of speciesi To make your calculations even easier, I’ve also posted (on the lab web site) a table that has the values for all of the (N log N) and (ni log nI) combinations of N or ni up to 490. Here is an example of how to calculate the Shannon-Wiener diversity index using method #2. Table 1.Species data from 5, 1/10 ha. plots at Otter Creek Natural Area Plot 1 Plot 2 Plot 3 Tree Species 1 Hemlock 12 Hemlocks 6 Hemlock 1 Chestnut Oak 1 Black Birch H’ = 0 0.221 0 Here’s how to use the rearranged equation and the (N log N) table So for plot 2 in the previous table 1) (N log N) = 16.046 N = 14 2) [n1 log n1] for hemlocks = 12.950 n1 = 12 3) [n2 log n2] for black birch = 0 n2 = 0 4) [n3 log n3] for chestnut oak = 0 n3 = 0 H’ = [(16.046) - Σ (12.95) + (0) + (0)]/14 → H’ = 0.221 Rev. 9/2005 Plot 4 3 Hemlock Plot 5 2 Hemlocks 2 Black Birch 0 0.301 BIO 300 7 Ecology Community Similarity Community ecologists have sought ways to express degree of similarity of communities to each other in composition and how this similarity relates to habitat conditions. They have invented a number of indices of similarity. We will use the simplest index, the coefficient of community similarity. Often it is useful to have a quantitative measure of the degree of similarity of two communities. The coefficient of community similarity is defined: CC = 2c/(s1 + s2) where c is the number of species in common between the two communities, s1 and s2 are the total number of species in each of the two communities. Note that when two communities have the same 10 species, CC = 20/(10 + 10) = 1. Today’s activities. Today’s lab will be based upon the following paper: Allan, J. David, Harvey J. Alexander, Ronald Greenberg. 1975. Foliage arthropod communities of crop and fallow fields. Oecologia 22:49-56. (on EReserves via the library website; the password is hosea) You will randomly sample and compare two areas for plant and arthropod diversity. 1) a goldenrod field with high structural diversity and low species richness and 2) a recently mowed field of foxtail (Setaria glauca) that has lower structural and but perhapsh greater species richness. 1) 2) There will be five teams of people. Each team will have separate self-selected people to conduct the foliage sampling and the arthropod sampling. Note: This lab does not require that you know all of the plant and insect species, but that you can reliably recognize and distinguish among different species. 1) 2) 3) 4) 5) 6) Using 2, 50 meter measuring tapes, you will set up a grid in both areas. Using a random numbers table, select coordinates and lay down the 1 meter square. Make 10 sweeps with the insect net. Place end of net in killing jar and loosely screw on lid. When insect have succumbed to the fumes, dump them out into the sorting tray. Sort by type and the number of each type that are similar. After the quadrat has been sampled for insects, the plant team can a. For each plant species you find, count and record the number of individuals. b.Estimate the proportion of foliage by area which has attained a height in one of three categories; < 15 cm, 15-45cm, >45 cm. 7) Sample 2 quadrats for each habitat. Rev. 9/2005 BIO 300 Ecology 8 We will combine data from the whole lab, therefore, it is up to you to coordinate amongst yourselves and agree on how you will designate each species of insect or plant. We will assign one person as curator for the insects and one as a curator for the plants who will then record a description and perhaps a name (e.g. black and yellow beetle, or bug species # 2). Individuals who ‘discover’ each new species may want to name their species. Calculations required 1. Generate a separate species area curve for the arthropods and plants for each habitat. a. I will assign each group a number, and you will plot the data from each group in numerical order to represent successive samples. 2. Generate a performance curve for the high diversity field. Calculate a mean density of Indian tobacco (Lobelia inflata) after each successive quadrat is sampled and plot those estimates (mean density on the y-axis, number of quadrats on x-axis.) Calculate each new mean using the numerical order of samples as in # 1. 3. Two-step sampling for Indian tobacco. Using N, x - the sample mean of Indian tobacco, and S the standard deviation, determine with a 90% confidence, whether your sample mean is within an interval of ± 10% of your mean. How many samples will it take to obtain a sample mean within 10% of the true mean? 4. Diversity and evenness calculations: a) Using the methods described by Allan et al. on page 2, calculate the mean foliage height diversity. To do so, calculate the foliage height diversity for each plot. (You will have 3 percentages for each plot. Be sure to convert these to decimal fractions.) Then average the foliage height diversity for the 10 quadrats. b) Calculate the mean Shannon-Weaver evenness index (for the plants sampled) using the equation on page 3 Allen et al.’s paper). This is not the Shannon-Weaver diversity index! The equation from the paper is : S - ∑ pi loge pi i =1 e= logeS Where S = the number of species. First – Calculate the top half of the equation. This is the formula for H’, the Shannon-Weaver diversity index that is on page 4-7 of this handout. If you are not a wiz with MS-Excel, then use the n log n table I’ve provided. (For the long form of the index, it does not matter what logarithmic base is used, as long as it is consistent. The table I supply for the shortcut method is base 10.) Second – After you have calculated the top half of the equation, divide by log10 S, where is S is the number of species. Note Since the shortcut uses base 10, then you should divide by log10 S. Your answer should be less than 1.0. 5. Calculate the mean Shannon-Weaver evenness index (as described above) for the arthropods sampled. Use the n log n table I’ve provided. 6. Calculate the community coefficient of similarity (page 7 of this handout) for both the plants and arthropods. Rev. 9/2005 BIO 300 Ecology 7. Calculate the ratio of number of predators for each habitat. Use the # of individuals, not the # of number of herbivores species. The Write-up - This is your first lab report for Ecology. Here’s what I want. 1) Write a short (1-2 paragraphs) introduction describing the 2 habitats and provide hypotheses regarding which habitat should have a greater species richness and species evenness of insects. Be sure to provide reasoning to support your selection of hypotheses. 2) No methods section. 3) In your results section, clearly present your a. Species area curve b. Performance curve for Indian tobacco. c. Your two-step sampling result for Indian tobacco.(i.e. your confidence interval) d. Mean Shannon-Weaver evenness index for foliage height diversity (both habitats). e. Mean Shannon-Weaver evenness index for plant species (both habitats). f. Mean Shannon-Weaver evenness index for arthropod species (both habitats). g. The community coefficient of similarity for plants and arthropods (to compare habitats). [For 3d, e, & f, calculate S-W evenness separately for each plot, then report mean + S.E. for each field.] 4) In your discussion, address the following; a. From your species area curve. Was sampling adequacy achieved for any of the organism/habitat combinations? If so, what is the sample size? What differences between the habitats might explain this? b. Is a stable mean obtained with the performance curve? c. From your two-step sampling, how many more samples would be required to obtain a population mean for Indian tobacco within a 10% interval with 90% confidence? Does your performance curve suggest that you were close to the population mean? d. Consider the results of Allan et al. In the second sentence of their discussion, they compare their results for crop and fallow fields. Fallow fields have: i. More species ii. Few individuals (per species) iii. Higher evenness iv. Proportionally more predator individuals. ⇒ Which of the fields you sampled corresponds to their crop field and which corresponds to their fallow old field? ⇒ Where did you find the greatest total number of insects, the mowed field of foxtail, or the goldenrod field? ⇒ How do your results compare to theirs? Elaborate. number of predators for each of the fields? Does it seem to be number of herbivores related to the species or structural diversity of the field? d. What is the ratio of Rev. 9/2005 9 BIO 300 10 Ecology DATA SHEET FOR SPECIES AREA CURVES AND HABITAT DIVERSITY PLOT NUMBER _____________ HABITAT TYPE __________________________ PLANTS Species ARTHROPODS Quantity (#) Species Quantity (#) ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ _____________________________ _______ Rev. 9/2005 BIO 300 11 Ecology DATA SHEET FOR SPECIES AREA CURVES AND HABITAT DIVERSITY PLOT NUMBER _____________ HABITAT TYPE __________________________ PLANTS Species ARTHROPODS Quantity (#) Species Quantity (#) ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ ________________________ ______ Rev. 9/2005