1.1.4. Inheritance and Recombination
advertisement
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
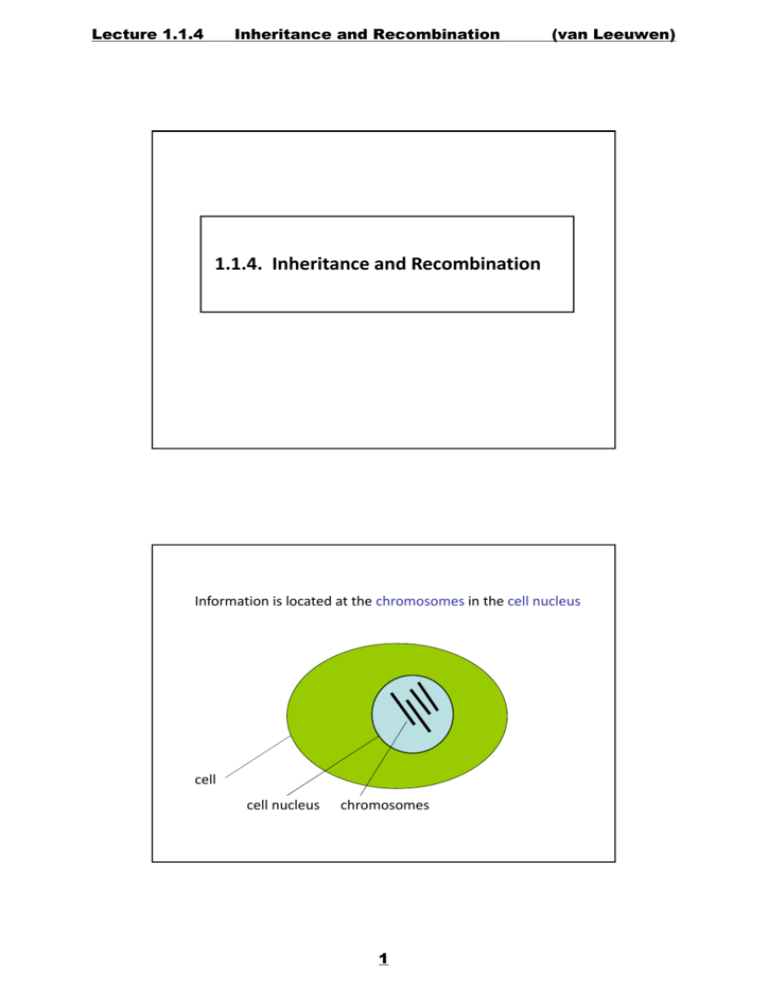
1.1.4. Inheritance and Recombination
Information is located at the chromosomes in the cell nucleus
cell
cell nucleus chromosomes
1
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
1933 Morgan ‐ Awarded Nobel Prize for theory of the gene genes
normal or branched bunch
round or ribbed fruit
Chromosome
of tomato
green or red stem colour
hairy or non‐hairy flower stem
resistance or susceptibility to Vertcillium
resistance or susceptibility to Fusarium
normal or pointed fruit
Mendel didn’t know the concept of a gene, but he mentioned it a character
Gene = Information on one special characteristic
One special plant species:
Special gene is always located at the same location at the chromosome
Content of information of one gene can differ (Mendel)
alleles A1 or A2 also sometimes written as alleles A or a
2
Lecture 1.1.4
Inheritance and Recombination
Chromosome 1 Chromosomes
Chromosome 1
(from ♀) (from ♂)
A
(van Leeuwen)
of tomato
a
2 homologous chromosomes
B
B
C
c
d
d
E
e
with genes on a fixed location (locus)
homozygous = AA or aa
A1A1 or A2A2
heterozygous = Aa
A1A2
Mendel:
F
f
If A be taken as denoting one of the two constant characters, for instance the dominant, a the recessive, and Aa the hybrid form in which both are conjoined, the expression
g
g
A + 2Aa + a
shows the terms in the series for the progeny of the hybrids of two differentiating characters.
two locules or many locules
normal or pear form
normal or necrotic
normal or oval
chromosome of tomato, each gene has 2 alleles
3
smooth or peach skin
normal or spotted
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
genetic information on genes on chromosomes
1 set of different chromosomes = genome
most species:
2 homologous chromosomes per chromosome = diploid (2n = ..)
in plant cell: 2 sets of chromosomes, 2 genomes
in gamete: 1 genome = haploid (n = ..)
polyploidy: more than two genomes (2n = 4x = ..)
allele for purple flowers
locus for flower‐
color gene
homologous pair of chromosomes
dipoid = 2n
allele for white flowers
A1A1
gametes: A1 A2A2
gametes: A2
A1A2
gametes: A1 and A2
haploid = n
4
Lecture 1.1.4
Inheritance and Recombination
Chromosome 1 Chromosomes
Chromosome 1
(from ♀) (from ♂)
of tomato
A
a
B
B
C
c
d
d
E
e
F
f
g
(van Leeuwen)
2 homologous chromosomes
with genes on a fixed location (locus)
homozygous = AA or aa
A1A1 or A2A2
heterozygous = Aa
A1A2
g
Parent 1
Parent 2
2n = 6
Meiosis =
Development of gametes
gamete
with meiosis the number of chromosomes is halved
gamete
n=3
in a gamete:
1 complete set of chromosomes
fertilisation
(1 genome)
zygote
2n = 6
5
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Intermediate inheritance
gametes
gametes
A1
A2
genotype: A1A1
red
A1A1
white
A2A2
phenotype: red flower
gametes
gametes
pink
A1A2
pink
A1A2
crossing A1A1 x A2A2
egg cells
pollen grains
½ A1
½ A2 ½ A1
¼ A1A1
¼ A1A2
½ A2
¼ A1A2
¼ A2A2
Punnett square
¼ A1A1 + ½ A1A2 + ¼ A2A2 genotype
¼ red + ½ pink + ¼ white
6
phenotype
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Dominant inheritance: PP and Pp have the same phenotype
X
parents: PP X pp purple white
F1: Pp purple selfing
F2
A Punnett square depicting a cross between two pea plants heterozygous for purple (B) and white (b) blossoms
7
Lecture 1.1.4
Inheritance and Recombination
Pp x Pp
Pp x pp
PP x pp
PP x Pp
(van Leeuwen)
1 PP + 2 Pp + 1 pp
genotype
3 purple + 1 white
phenotype
1 Pp + 1 pp
genotype
1 purple + 1 white
phenotype
1 Pp
genotype
1 purple
phenotype
1 Pp + 1 PP
1 purple
genotype
Difference between BB and Bb
•Selfing
BB BB
Bb BB, Bb and bb
•Backcrossing with bb
BB x bb Bb
Bb x bb Bb and bb
•Molecular markers can see the difference between BB and Bb
8
phenotype
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Reciprocal crossing
•aa x AA Aa
selfing aa
see the difference
aa
•AA x aa
Aa
selfing AA
AA
don’t see the
difference
Overdominance
Aa is more than AA, is more than aa
Aa > AA > aa
Dominance ratio’s
aa
aa
Aa
AA
intermediate
AA
dominance
Aa
aa
AA
9
Aa
overdominance
Lecture 1.1.4
Inheritance and Recombination
Codominance
Both alleles are active and code for different ‘actions’ :
Incompatibility (no fertilization):
S1S1
prohibits S1 S1S2
prohibits S1 and S2 S2S2
prohibits S2 meiosis
B
A
BB
a
b
A
or
b
a
2 AB and 2 ab or 2 Ab and 2 aB
10
(van Leeuwen)
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
With dominance, what are different phenotypes?
AaBb
Egg cells
¼ AB + ¼ Ab + ¼ aB + 1/4 ab
1/4 AB
1/4 Ab
1/4 aB
1/4 ab
1/4 Ab
AABb
AAbb
AaBb
Aabb 1/4 aB
AaBB
AaBb aaBB
aaBb
1/4 ab
AaBb
Aabb
aaBb
aabb
Pollen
1/4 AB
parents
gametes
egg cells
pollen grains
11
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
A/a: colour
B/b: length
white, long x purple, short
F1 = purple, long
genotype parents and F1?
F2?
white, long x purple, short F1 = purple, long gametes:
F2
phenotype
genotype
12
Lecture 1.1.4
Inheritance and Recombination
A1/A2: colour
B/b: wide/narrow
(van Leeuwen)
genotype parents and F1?
gametes F1?
inheritance?
genotype and phenotype F2?
white, wide x red, narrow F1 = pink, wide
F2
phenotype
genotype
13
gametes:
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
fem. gametes
m. gametes
¼ AB
¼ Ab
¼ aB
¼ AB
¼ Ab
¼ aB
¼ ab
¼ ab
A1A1BB
A1A1Bb
A1A2BB
A1A2Bb
A1A1Bb
A1A1bb
A1A2Bb
A1A2bb
A1A2BB
A1A2Bb
A2A2BB
A2A2Bb
A1A2Bb
A1A2bb
A2A2Bb
A2A2bb
2 methods of determining the progeny:
1. via a punnett scheme
2. Take first the segregation of gene A/a and than gene B/b
A1A2Bb (pink)
¼ A1A1 (red) ¾ B. (wide) 3/16 A1A1B. (red, wide)
¼ bb (narrow) 1/16 A1A1bb (red, narrow
½ A1A2 (pink)
¾ B. (wide) 6/16 A1A2B. (pink, wide)
¼ bb (narrow) 2/16 A1A2bb (pink, narrow)
¼ A2A2 (white)
¾ B. (wide) 3/16 A2A2B. (white, wide)
¼ bb (narrow) 1/16 A2A2bb (white, narrow)
Colour is intermediair, Wide dominant
Segregation with 3 genes (all dominant inheritance)
segregation
for A
segregation
for A and B
segregation 27/64
for A, B and A.B.C.
C
3/4
A.
1/4
aa
9/16
A.B.
A.B.cc
3/16
A.bb
A.bbC.
A.bbcc
14
3/16
aaB.
1/16
aabb
3/64
aabbC.
1/64
aabbcc
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
General scheme for independent Mendel segregation
starting from heterozygous F1 and selfing it.
Number
of
genes
number of
different
gametes
number of
number of
squares in the genotypes
Punnett
in F2
scheme
number of
phenotypes
in F2
(dominance
number of
phenotypes in
F2
(intermediate
2
22 = 4
(22)2 = 16
32 = 9
22 = 4
32 = 9
3
23 = 8
(23)2 = 64
33 = 27
23 = 8
33 = 27
4
24 = 16
(24)2 = 256
34 = 81
24 = 16
34 = 81
1
n
AaBBCc x aaBbCc, which is the fraction of aaBBCC in the progeny ??
15
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
If we have deviations in segregation numbers from the expected ratio, when is that deviation so large that we are going to doubt our assumption about the inheritance?
For example: we think it is intermediate inheritance and we expect a ratio of ¼ AA + ½ Aa + ¼ aa
So if we have 64 seeds from a selfing of Aa we expect: 16 AA + 32 Aa + 16 aa
The difference between the observed value from the expected value is determining if the deviation is reasonable (due to coincidence) or is so large that the deviation is not coincidental,
something else is the case (wrong expectation, due to wrongly determination of inheritance, mistakes in observation, and so on)
Expected:
16 AA + 32 Aa + 16 aa
Observed: 18 AA + 32 Aa + 14 aa
So the difference between xobs en xexp is the measure for our judgement
16
Lecture 1.1.4
Inheritance and Recombination
2 df i
( xobs. xexp. ) 2
xexp.
df means degrees of freedom that is number of different phenotypes ‐ 1
The larger the difference, the larger the value of Χ2
Expected:
16 AA + 32 Aa + 16 aa
Observed: 18 AA + 32 Aa + 14 aa
Χ2 =
df =
17
(van Leeuwen)
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
In the table:
Χ2 =
df =
Chance P (horizontally) with differen t degree s of freedom (df),
that such a large deviation is c oincidental.
P 0,90
df
1
2
3
0,70
0.50
0,30
0,20
0,10
0,05
0,01
0,016 0,15
0,21 0,71
0,58 1,42
0,46
1,39
2,37
1,07
2,41
3,67
1,64
3,22
4,64
2,71
4,61
6,25
3,84
5,99
7,82
6,64
9,21
11,35
Linkage
Linkage means that two genes are located at the same chromosome
Linked genes stay together in meiosis
Unless the linkage is broken by crossing over
Crossing over
18
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
meiosis causes genetic variation by:
1. different combination of chromosomes (and genes)
2. new combinations of genes by crossing over
No crossing over between A/a and B/b
A
a
A
A
a
a
B
b
B
B
b
b
AB
AB
ab
ab
Crossing over
A
a
A
A a
a
A
A
a
a
B
b
B
b B
b
B
b
B
b
aB
ab
AB
19
Ab
Lecture 1.1.4
Inheritance and Recombination
Crossing over
diploid cell, 2n=6
AaBbCc
A a (van Leeuwen)
ABCD ABCd
meiose
B b abcD abcd
c D d
C A C
B D
A C
a
B
d a
b D
c b c d
Plant: AaBb
A
B
coupling phase: dominant allele linked to dominant allele
a
b
80% of the gametes are formed with crossing over
20% of the gametes are formed without crossing over
Think of 10 gamete forming cells
Each of these cells produce 4 gametes
cell with crossing over ‐‐‐> gametes AB, Ab, aB and ab
cell without crossing over
‐‐‐> gametes 2 AB and 2 ab
20
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
10 cells in meiosis form 40 gametes
A
B
a
b
Plant AaBb
8 with and 2 without crossing over
AB
Ab
aB
A
C
a
c
Ac
aC
ab total
8 with crossing over
2 without crossing over
total
% recombinants =
10 cells in meiosis form 40 gametes
Plant AaCc
2 with and 8 without crossing over
AC
2 with crossing over
8 without crossing over
total
recombinants =
21
ac totaal
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Plant: AaBb
A
repulsion phase: dominant allele linked to recessive allele
b
B
a
20% of the gametes are formed with crossing over
80% of the gametes are formed without crossing over
Think of 10 gamete forming cells
Each of these cells produce 4 gametes
cell with crossing over ‐‐‐> gametes AB, Ab, aB and ab
cell without crossing over
‐‐‐> gametes 2 Ab and 2 aB
10 cells in meiosis form 40 gametes
A
d
a
D
Plant AaDd
9 with and 1 without crossing over
AD
Ad
aD
ad total
9 with crossing over
1 without crossing over
totaal
recombinants =
p = r = recombinant fraction = measure for genetic distance
22
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Relative frequencies of the gametes in a formula with recombination fraction r (grey colour are recombinants).
gametes
AB
Ab
aB
ab
Total
½ (1‐r)
1
½ r
1
Coupling phase
½ (1‐r)
½ r
½ r
Repulsion phase
½ r
½ (1‐r)
½ (1‐r)
r=0
r = ½.
Absolute linkage
No linkage
r is a measure for the distance between 2 loci.
AaBb x aabb
gametes AB, Ab, aB, ab
ab
no linkage
linkage
AaBb
250
300
Aabb
250
200
aaBb
250
200
aabb
250
300
23
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
P1 SSRR
x P2 ssrr
leaves with a stem
leaves without stem, flowers red flowers pink
F1 = 100% SsRr
leaves with stem, red flower
r = ?
r =
The crossing F1 x P2 gives the following result:
gametes
F1
gamete P2 sr
phenotype
number
SR
SsRr
with stem, red flower
94
Sr
Ssrr
with stem, pink flower
6
sR
ssRr
without stem, red flower
sr
ssrr
without stem, pink flower
8
92
Division of gametes which are formed by the triheterozygous plant AaBbCc in 4 situations Gametes 1 2 3 4 AaBbCc ABC 100
150
300
ABc 100
150
10
AbC 100
50
50
Abc 100
50
40
aBC 100
50
40
aBc 100
50
50
abC 100
150
10
abc 100
150
300
800
800
800
24
400 0 0 0 0 0 0 400 800 Lecture 1.1.4
Inheritance and Recombination
situation 3:
AB
Ab
aB
ab
AC
Ac
aC
ac
BC
Bc
bC
bc
(van Leeuwen)
rab =
rac =
rbc =
situation 3
0,225
A
C
0,125
B
0,15
0,225 < 0,125 + 0,15 !!
double crossing over
25
Lecture 1.1.4
Inheritance and Recombination
A
C
(van Leeuwen)
B
A
C
B
a
c
b
a
c
b
A C B
ACB
A c B
AcB
aCb
a C b
a c b
double crossing over
ACB
AcB
=10/800 = 0,0125
aCb
=10/800 = 0,0125
acb
+ 0,025
This influences both the combination A/C and B/c: 2 x 0,025 = 0,05
0,275 – 0,225 = 0,05
26
acb
Lecture 1.1.4
Inheritance and Recombination
situation 2:
AB
Ab
aB
ab
rab =
AC
Ac
aC
ac
rac =
BC
Bc
bC
bc
rbc =
Conclusion?
Genes on chromosomes
genetic map of chromosome 1 of tomato
genetic map of chromosome 9 of mais
27
(van Leeuwen)
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Linkage causes different segregation ratio’s Linkage: if ‘good’ genes are linked it is favourable and it can be used for indirect selection
gene and marker can also be linked
Linkage: if a ‘good’ gene is linked to a ‘bad’ gene it is unfavourable and recombination is necessary
pleiotrophy: one gene is influencing two different characters
very close linkage can only be discriminated from pleiotrophy if you find the recombinant
examples:
cucumber: resistance gene for powdery mildew with necrosis
2 resistance genes for downy mildew in spinach in repulsion phase
example of a rare recombinant:
lettuce: Nasinovia (aphid) resistance and dwarf growth
28
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Epistasis
Epistasis:
2 genes influence one charcter
and influence sometimes each other expression
parents
F1
F2
2 genes, 2 characters
29
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
F2 of AaBb with double dominance:
4 phenotypes, 1 character
3 A.
1 aa
3 B.
9 A.B.
3 aaB.
1 bb
3. A. bb
1 aabb
Epistasis: 1 gene influences the expression of the other gene
Genes A and B influence the same character,
but don’t influence each other expression:
e.g. 4 different colours
3/4 A.
1/4 aa
3/4 B.
9/16 A.B.
3/16 aaB.
1/4 bb
3/16 A.bb
1/16 aabb
gene pairs A/a
B/b
30
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Parental cross
F1
F2
1 single
9 walnut 3 rose 3 pea
genotypes?
Genes Y/y and Cl/cl influence the same character,
but don’t influence each other expression:
e.g. 4 different fruit colours in pepper
gene pairs Cl/cl
3/4 Cl.
1/4 clcl
3/4 Y.
9/16 Y.Cl.
3/16 Y.clcl
1/4 yy
3/16 yyCl.
1/16 yyclcl
Y/y
31
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
One gene influences the expression of the second gene = epistasis
If A is dominant, you don’t see the difference between B. and bb Dominant epistasis
3/4 A.
1/4 aa
3/4 B.
9/16 A.B. +
3/16 aaB.
1/4 bb
3/16 A.bb = 12
1/16 aabb
gene pairs A/a
B/b
F2
12/16 white
32
Lecture 1.1.4
Inheritance and Recombination
recessive epistasis
If a is recessive, you don’t see a difference between B. and bb
genenparen A/a
B/b
3/4 A.
1/4 aa
3/4 B.
9/16 A.B.
3/16 aaB. +
1/4 bb
3/16 A.bb
1/16 aabb = 4
33
(van Leeuwen)
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
flower colour of beans
A. =anthocyane
P1 aaBB x white P2 AAbb
red
aa = no anthocyane
B. = basic cell plasma
F1 = AaBb
purple
bb= acid cell plasma
F2 = 9/16 A.B. + 3/16 A.bb + purple red 9 : 3 : 34
3/16 aaB. + 1/16 aabb
white white
4 (= 3 + 1)
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Reciprocal dominant epistasis
If at least one of the genes is dominant, there is an effect,
only aabb is different
Example: Powdery mildew in Cucumber.
Only the recessive genotype pm1 pm1 pm2 pm2 pm3 pm3 is resistant. All other genotypes are susceptible
genenparen A/a
B/b
3/4 A.
1/4 aa
3/4 B.
9/16 A.B.
3/16 aaB.
1/4 bb
3/16 A.bb
1/16 aabb
recombination
•crossings
combination of good characteristics from 2 or more varieties or 2 lines
•repeated backcross
combination of variety with 1 good characteristic from a wild species
•selfing
recombination of the genes of a heterozygous plant
•doubled haploids
recombination of the genes of a heterozygous plant
•transformation/genetic modification
combination of a variety with a piece of useful DNA
•mutation
1 gene is mutated (spontaneously or by treatment)
35
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Sexual propagation means meiosis, in order to develop gametes
Meiosis is the first recombination moment, when making a crossing, a back‐crossing, a selfing or doubled haploids
Recombination events in a breeding program:
•crossing to increase genetic variation
•F1 hybrid seed production crossing
•back crossing
•selfing
•doubled haploids (not in each crop)
AAbbCCdd x aaBBccDD
AaBbCcDd
AABbCCdd
aaBbCCdd
AaBbCCDD
AAbbCcDD
etc.
If crossing parents are homozygous segregation in F2
If crossing parents are heterozygous segregation in F1
36
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Parents are more or less heterozygous if:
•Open Pollinated varieties
•Lines of some crops are not homozygous due to inbred depression
•Vegetatively propagated crops
•Use of hybrids as crossing parents
•Use F1 to cross with a third parent
X
Segregation in F1
Combination of 2 lines differing in 5 monogenic traits in order to make the perfect plant with these 5 traits
AaBbCcDdEe selfing
What is the expected fraction of AAbbCCDDee ?
=
• can you recognize the correct phenotype?
• dominance of genes? (AA or Aa)
• still available also in heterozygous form (e.g. AaBbCcDdEe)
• size of F2?
37
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Repeated back crossing
After crossing with wild species:
Characteristics of modern variety have to be regained by repeated back crossing
The F1 is crossed again with modern variety back to
Repeated back crossing
Variety x Wild
x
F1 x Variety
1/2 wild
x
BC1 x Variety
1/4 wild
x
1/8 wild
x
BC2 x Variety
38
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
repeated backcrossing and introgression
Introgression lines
39
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Different genotypes after 5 generations of backcrossing
Repeated backcrossing is also used for:
•transferring a gene from one type to another type within the species
•transferring a gene from a competitors variety to your own variety
•to introduce male sterility in a line
40
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
How many times backcrossing?
Depends on: •How wild is wild?
•Linkage desired characteristics with undesirable wild trait
•Crossing over close to the desired gene
Chromosome with desired trait
Desired trait
Wild Variety BC1
BC2
General cultural value
Cultivar
Modern variety
wild
Wild species
F1 BC1 BC2 BC3 BC4 BC5
41
BC3 BC4
Lecture 1.1.4
P1
Inheritance and Recombination
(van Leeuwen)
BC1 population
P2
Parental generation
x
F1
P2
Gametes of
P2
x
Gametes of
F1
BC1 generation
which plant has the most ‘cultivar’like genome?
visual selection or molecular markers
Selfing
Selfing:
natural method of reproduction for self pollinators
Selfing with cross pollinators:
(for making inbred lines)
Covering or isolating plants
result of selfing is the same as with self pollinators
42
Lecture 1.1.4
Inheritance and Recombination
Generation Segregation of genotypes
AA
0
0,25
F1
F2
F3
F4
F5
F6
F7
F8
Aa
1
0,5
(van Leeuwen)
%
heterozygotes
aa
0
0,25
100%
50%
F∞
Self fertilization:
Increasing of the number of homozygous plants
result: 2 homozygous genotypes
P1
F2 population
P2
Parental generation
x
Parents homozygous
Selfing is possible
F1
F1
x
F1 generation
Gametes of
F1 generation
F2 generation
43
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Selfing of AaBbCc
Which homozygous genotypes?
AABBCC, AAbbCC, aaBBCC, aabbcc
AABBcc, AAbbcc, aaBBcc, aabbcc
nr. different homozygous genotypes ?
With 10 genes: nr. different homozygous genotypes ?
General: 2n different homozygous genotypes
n = number of different genes, which are heterozygous in F1
Selfing of AaBbCc
Which homozygous genotypes in F2?
Which fraction of genotypes in F2 is homozygous?
Totally 8/64 = 1/8 homozygous = ( ½ )3
General : ( ½ )n n=10 ( ½ )10 = 1/1024 is homozygous
So also in next generations (F3 – F5) there is a lot of recombinations, as there are still a large number of heterozygous plants
44
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
% hom ozygoten
120
100
80
60
40
20
0
1
2
3
4
5
6
7
8
9 10 11 12 13
Generatie (F)
Increasing numbers of homozygotes with self fertilization
for 1 gene (purple) and 20 genes (yellow)
Remarks:
Genetic variation in F2 and later generations dependent on genetic distance between crossing parents
In F2 not all desirable combinations are available, but they may appear in later generations
F2 as large as possible (size limited by space and labour)
F7‐F8 is considered to be homozygous:
In reality still segregation possible
45
Lecture 1.1.4
Inheritance and Recombination
(van Leeuwen)
Doubled haploids
•From different gene pools two lines are crossed
•The F1 is heterozygous (AaBbCcDDEeFF)
•In the very young flowers gametes develop by meiosis
(a lot of diffent gamets, e.g. ABCDeF)
microspores or macrospores (not yet fully developed pollen grains or egg cells) •Genome is doubled spontaneously or by colchicine
•Embryo grows on tissue culture, untill it is big enough to grow in a pot
doubled haploids
AaBbCcDd
Plant
2n
meiosis
abcD
aabbccDD
Abcd
AAbbccdd
AbCD
AAbbCCDD
ABCd
AABBCCdd
aBCd
aaBBCCdd
Gametes
n
colchicine
Homozygous plants
2n
Recombination
46
Lecture 1.1.4
Inheritance and Recombination
Doubled haploids, microspore culture
Pollen mother cell (PMC)
Development in vitro
Ripe pollen grain
•doubled haploids
from microspores/anthers: pepper, cabbage
from egg cells: cucumber, maize
47
(van Leeuwen)
Lecture1.1.5
Polulation genetics
1.1.5. Population genetics
A population is a group of individuals
In the genetic sense, it is a breeding group, consisting of individuals
population is characterised by 1. the genotypes of the individuals
2. by the transmission of genotypes to the next generation
48
van Leeuwen
Lecture1.1.5
Polulation genetics
van Leeuwen
What is a genotype?
The combination of alleles located on homologous chromosomes that determines a specific characteristic or trait.
Cells Chromosomes DNA Gene
generative propagation via gametes
stigma receives pollen
antheres produce pollen grains (male gametes)
ovules produce female gametes
selfing or outcrossing has influence on the transmission of genes cross pollination to the next generation
49
Lecture1.1.5
Polulation genetics
van Leeuwen
population consists of plants with different genotypes
A1A1 and A2A2 and A1A2
plant
egg cells
pollen grain
A1A1
A1
A1
A2A2
A2
A2
A1A2
A1 + A2
A1 + A2
population of 100 plants
plant
number of individuals
Number of genes A1
Number of genes A2
A1A1
30 A1A2
A2A2
60 10
Total
100
60 60 0 120
200
0 60 20 80
50
Lecture1.1.5
Polulation genetics
A1A1
Number of genotypes
Frequencies
Genotypes
A1A2
van Leeuwen
A2A2
Total
30
0,3
60
0,6
10
0,1
100
1
Freq. Falconer
P
H
Q
1
Freq. Bernardo
P11
P12
P22
1
Genotype frequencies
population of 100 plants
plant
A1A1
number of individuals
30 Number of genes A1
60 60 0 120
Number of genes A2
0 60 20 80
Gene frequency
symbols
Falconer
Bernardo
A1A2
60 A2A2
Total
10
100
A1
A2
(2*30+60)/200=0,6
(2*10+60)/200=0,4
p
q
P + ½ H
P11 + ½ P12
Q + ½ H
P22 + ½ P12
51
200
Lecture1.1.5
Polulation genetics
A1
p
Frequencies
P + ½ H Genes
A2
q
Total
1
A1A1
P
van Leeuwen
Genotypes
A1A2 A2A2 Total
H
Q
1
Q + ½ H
P11 + ½ P12 P22 + ½ P12
P11
P12
P22
populations can differ in:
•genotype frequencies and
•gene frequencies
Gene and genotype frequencies can be influenced by:
1. Population size (in small population there is influence of sampling variation)
2. Differences of fertility and viability (selection)
3. Migration
4. Mutation
5. Mating system
52
1
Lecture1.1.5
Polulation genetics
van Leeuwen
We start with a large population, with equal fertility and viability for all genotypes
with no migration and mutation
with a random mating system
For this population the Hardy Weinberg law is valid:
gene and genotype frequencies are constant from generation to generation
Sometimes a starting population is not in Hardy Weinberg equilibrium:
but after one round of random mating it is in HW‐equilibrium
population of 100 plants
plant
number of individuals
frequency of individuals
genes
gene
frequencies
A1A1
30 A1A2
A2A2
60 10
Total
100
P = 0,3 H = 0,6 Q = 0,1 1
A1
p = 0,6
A2
q = 0,4
53
Lecture1.1.5
Polulation genetics
van Leeuwen
Next generation:
gametes
p A1
p A1
q A2
p2 A1A1
pq A1A2
q A2
pq A1A2
q2 A2A2
p2 A1A1 + 2 pq A1A2 + q2 A2A2
Next generation:
gametes
0,6 A1
0,4 A2
0,6 A1
0,36 A1A1
0,24 A1A2
0,4 A2
0,24 A1A2
0,16 A2A2
If we calculate the gene frequencies of this population:
p =
the gene frequencies have not changed next generation: same genotype
frequencies
q =
Hardy Weinberg equilibrium
54
Lecture1.1.5
Polulation genetics
van Leeuwen
So in following generations:
A1A1
A1A2
A2A2
Total
Number of genotypes
30
60
10
100
Frequencies
0,3
0,6
0,1
1
Genes
A1
A2
0,6
0,4
A1A1
A1A2
A2A2
Total
0,36
0,48
0,16
1
Generation 0 Genotypes
Generation 1 Genotypes
Frequencies
Hardy Weinberg equilibrium
Number of genotypes
36 48 16 100
Genes
A1
A2
A1A1
A1A2
Generation 2 Genotypes
A2A2
Total
Frequencies
Number of genotypes
So if HW‐equilibrium has to be proved, take the following steps:
1. genotype frequencies in parents gene frequencies in gametes
gene frequencies in gametes forming zygotes
2. genotype frequencies in zygotes
3. genotype frequencies in parents gene frequencies in gametes
genotype frequencies in progeny
4. gene frequencies in progeny
HW‐equilibrium gene freq. in step 4 = gene freq. in step 3
or in step 3 genotype freq. of parents = genotype freq. progeny
55
Lecture1.1.5
Polulation genetics
van Leeuwen
Conditions:
1. Normal gene segregation
2. Equal fertility of parents
3. Equal fertilizing capacity of gametes
4. Large population
5. Random mating
6. Equal gene frequencies in male and female parents
7. Equal viability
8. No migration
9. No mutation
q
from:
Falconer&Mackay, 1996
56
Lecture1.1.5
Polulation genetics
model
Genes
A2
q
A1
p
Frequencies
Total
1
A1A1
P
van Leeuwen
Genotypes
A1A2 A2A2
H
Q
Total
1
Considering HW from the viewpoint of genotypes mating with each other
9 combinations
♀
♂
A1A1
P
A1A1
A1A2
A2A2
P
H
Q
P2
PH
PQ
HQ
Q2
A1A2
H
PH
H2
A2A2
Q
PQ
HQ
Genotype and frequency of progeny
Mating
Frequency
A1A1 x A1A1
P2
A1A1 x A1A2
2PH
A1A1 x A2A2
2PQ
A1A2 x A1A2
H2
A1A2 x A2A2
2HQ
A2A2 x A2A2
Q2
A1A1
A1A2
total
57
A2A2
Lecture1.1.5
Polulation genetics
van Leeuwen
human beings are outcrossing
rare genetic deviations are often recessive
and aa‐genotypes occur seldom
AA:
healthy
Aa:
healthy and carrier
aa:
diseased
If in a population of 16 million, there are 1600 persons having a certain disease (genotype aa),
how many persons are carrier (genotype Aa)? (Assuming random mating)
1600 in 16 million aa
q2 = ? A
p=
Male
a
q=
Female
1600 in 16 million aa
q2 =
A
p=
p2=
pq=
q = p = a
q=
pq=
q2=
healthy AA
healthy and carrier Aa
diseased aa
assumptions?
58
total
16 million
Lecture1.1.5
Polulation genetics
van Leeuwen
More than one locus, e.g. mixing of 2 populations A1A1B1B1
x
A2A2 B2B2
→
A1A2 B1B2
↓
random mating of A1A2 B1B2
frequency
A1A1B1B1
A1A2B1B1
pA =
A2A2B1B1
qA =
A1A2 B1B2
A1A1B1B2
pB =
gametes
qB =
A2A2B1B2
A1A1 B2B2
A1A2 B2B2
A2A2 B2B2
assumptions: no linkage
A1B1 +
A1B2 +
A2B1 + A2B2
More than one locus, e.g. mixing of 2 populations (equal size), 3 possible combinations with random mating A1A1B1B1
x
A2A2 B2B2
→
A1A2 B1B2
0,5
A1A1B1B1
x
A1A1 B1B1
→
A1A1 B1B1
0,25
A2A2B2B2
x
A2A2 B2B2
→
A2A2 B2B2
0,25
generation 0
random mating gametes A1B1 and A2B2
other gametic types and combinations of genotypes occur and increase in coming generations:
at this stage: linkage disequillibrium
(irrespective if the loci are linked)
in case of linkage the approach to equilibrium will take more time
59
Lecture1.1.5
Polulation genetics
van Leeuwen
If in equilibrium (after a number of generations random mating):
Genes
Gene frequencies
Gametic types
Frequencies, equilibrium
Frequencies, actual
Difference from equilibrium
A1
pA
A2 qA B1 pB B2 qB 0,5
A1B1 pA pB
0,5
A1 B2 pA qB 0,5
A2 B1 qA pB 0,5
A2 B2 qA qB 0,25
0
0,25
0
0,25
0
0,25
0
If not in equilibrium yet (after random mating):
Genes
Gene frequencies
Gametic types
Frequencies, equilibrium
Frequencies, actual
Difference from equilibrium
A1
pA
0,5
A1B1 pA pB
pA1B1
D
D = pA1B1 ‐ pA1 * pB1
60
A2 qA B1 pB B2 qB 0,5
0,5
0,5
A1 B2 A2 B1 A2 B2 pA qB qA pB qA qB pA1B2 pA2B1 pA2B2
‐D
‐D
D
Lecture1.1.5
A1B1
A1B2
A2B1
A2B2
Polulation genetics
A1A1B1B1 0,25
0,25
A1A2 B1B2 0,5
0,125
0,125
0,125
0,125
A2A2 B2B2 0,25
0,25
van Leeuwen
gametes
0,375
0,125
0,125
0,375
Development towards equilibrium (after 1 generation of random mating):
Genes
Gene frequencies
Gametic types
Frequencies, equilibrium
Frequencies, actual
A1
pA
0,5
A2 qA 0,5
B1 pB 0,5
B2 qB 0,5
A1B1 0,25
A1 B2 0,25
A2 B1 0,25
A2 B2 0,25
0,375
Difference from equilibrium
D=
61
0,125
0,125
0,375
Lecture1.1.5
Polulation genetics
van Leeuwen
Developing towards an equilibrium (after a number of generations random mating):
A1B1 0,25
A1 B2 0,25
A2 B1 0,25
A2 B2 0,25
0,375
0,125
0,125
0,375
Frequencies after 2 gen. rm
0,3125
0,1875
0,1875
0,3125
Frequencies after 3 gen. rm
0,28125 0,21875 0,21875 0,28125 0,03125
Frequencies after 4 gen. rm
0,26562 0,23437 0,23437 0,26562 0,00781
Frequencies after 5 gen. rm
0,25781 0,24219 0,24219 0,25781 0,00781
Gametic types
Frequencies, equilibrium
Frequencies after 1 rm
D
0,25
0,125
0,0625
Disequilibrium, because of starting with a mix of two populations which are completely different (but no linkage): A1A1B1B1
and
A2A2 B2B2
Starting with the F1 heterozygous plant in coupling phase: A1B1/ A2B2 (so with linkage)
Genes
A1
A2 B1 B2 Gene frequencies symbols
pA
qA pB qB Gene frequency
0,5
0,5
0,5
0,5
Gametic types
A1B1 A1 B2 A2 B1 A2 B2 Frequencies in equilibrium
pA pB
pA qB
qA pB qA qB Frequencies, actual
Difference from equilibrium (D)
pA1B1= ½(1‐r) pA1B2= ½(r) pA2B1= ½(r) pA2B2= ½(1‐r) D= ¼ ‐ ½ r
½ r– ¼ =‐ D
D = pAiBj ‐ pAi * pBj
62
‐D
D= ¼ ‐ ½ r
Lecture1.1.5
Polulation genetics
van Leeuwen
gametes
A1 B2 A2 B1 r
F1: A1A2 B1B2 (coupling
phase)
A1B1 0,1
0,45
0,5*(1‐r)
0,05
0,5 r
0,05
0,5 r
0,45
0,5*(1‐r)
F2
A1A1B1B1 0,2025
A1B1 0,2025
A1 B2 A2 B1 A2 B2 A1A2B1B1 0,045
0,0225
A2A2B1B1 A1A1B1B2
A1A2 B1B2 (c)
A1A2 B1B2 (r)
A2A2B1B2
A1A1 B2B2 A1A2 B2B2 A2A2 B2B2 0,0025
0,045
0,405
0,005
0,045
0,0025
0,045
0,2025
0,0025
0,0225 0,0225
0,18225 0,02025 0,02025 0,18225
0,00025 0,00225 0,00225 0,00025
0,0225 0,0225
0,0025
0,0225
0,0225
0,2025
gametes F2
0,43
D0 = 0,45 – 0,52 = 0,2
F2 gametes
r =
A2 B2 0,0225
0,07
0,07
0,43
D1 = 0,43 – 0,52 = 0,18
0,1
A1B1 0,43
A1 B2 0,07
A2 B1 0,07
A2 B2 0,43
F3 random A1A1B1B1 mating
0,1849
A1B1 0,1849
A1 B2 A2 B1 A2 B2 A1A2B1B1 0,0602
0,0301
A2A2B1B1 A1A1B1B2
A1A2 B1B2 (c)
A1A2 B1B2 (r)
A2A2B1B2
A1A1 B2B2 A1A2 B2B2 A2A2 B2B2 gametes
0,0049
0,0602
0,3698
0,0098
0,0602
0,0049
0,0602
0,1849
0,0301
0,0049
0,0301
0,16641
0,00049
0,0301
0,01849
0,00441
0,01849 0,16641
0,00441 0,00049
0,0301 0,0301
0,0049
0,0301
0,412
D2 = 0,412 – 0,52 = 0,162
63
0,088
0,088
0,0301
0,1849
0,412
Lecture1.1.5
Polulation genetics
van Leeuwen
r
A1B1 A1 B2 A2 B1 A2 B2 D
0,1
0,45
0,05
0,05
0,45
0,2
F2
0,43
0,07
0,07
0,43
0,18
F3 (random mating)
0,412
0,088
0,088
0,412
0,162
F1
D0 = 0,2
D1 = 0,18 = D0 (1‐r) = 0,2 * (0,9)
Dt = D0 (1‐r)t
D2 =
1. gamete A1B1 can be produced as a non‐recombinant from genotype A1B1/AxBx
frequency: pA1B1 (1‐r) (r = recombination frequency)
2. gamete A1B1 can be produced as a recombinant from genotype A1Bx/AxB1
frequency: pA pX r (r = recombination frequency)
pA1B1’ = pA1B1 (1‐r) + pApB r
D’ = PA1B1’ - pA pB = pA1B1(1‐r) + pA pB r - pA pB
= (pA1B1 ‐ pA pB ) (1-r) = D (1-r)
64
Lecture1.1.5
Polulation genetics
F1
r
0,4
F2
A1A1B1B1 A1A2B1B1 A2A2B1B1 A1A1B1B2
A1A2 B1B2 (c)
A1A2 B1B2 (r)
A2A2B1B2
A1A1 B2B2 A1A2 B2B2 A2A2 B2B2 0,09
0,12
0,04
0,12
0,18
0,08
0,12
0,04
0,12
0,09
gametes
A1B1 0,3
0,5*(1‐r)
van Leeuwen
A1 B2 0,2
0,5 r
A2 B1 0,2
0,5 r
A2 B2 0,3
0,5*(1‐r)
A1B1 0,09
0,06
A1 B2 A2 B1 A2 B2 0,06
0,054
0,016
0,06
0,036
0,024
0,06
0,04
0,036
0,024
0,06
0,04
0,06
gametes F2
0,28
D0 =
0,22
0,054
0,016
0,06
0,06
0,09
0,22
0,28
D1 =
F3 random r =
mating
0,4
A1B1 0,28
A1 B2 0,22
A2 B1 0,22
A2 B2 0,28
F3 random A1A1B1B1 A1A2B1B1 A2A2B1B1 A1A1B1B2
A1A2 B1B2 (c)
A1A2 B1B2 (r)
A2A2B1B2
A1A1 B2B2 A1A2 B2B2 A2A2 B2B2 gametes
mating
0,0784
0,1232
0,0484
0,1232
0,1568
0,0968
0,1232
0,0484
0,1232
0,0784
F3
A1B1 0,0784
0,0616
A1 B2 A2 B1 A2 B2 0,0616
0,04704
0,01936
0,0616
0,03136
0,02904
0,0616
0,0484
0,03136 0,04704
0,02904 0,01936
0,0616 0,0616
0,0484
0,0616
0,268
D2 =
65
0,232
0,232
0,0616
0,0784
0,268
Lecture1.1.5
Polulation genetics
F1
F2
F3 (random mating)
F1
F2
F3 (random mating)
r
0,1
D
0,2
0,18
0,162
D0 = 1
1
0,9
0,81
r
0,4
D
0,05
0,03
0,018
D0 = 1
1
0,6
0,36
If r is low (narrow linkage) it takes more time before D = 0
Genes
Gene frequencies
Gametic types
Frequencies, equilibrium
Frequencies, actual
Difference from equilibrium
A1
pA
A1B1 pA pB
pA1B1
van Leeuwen
Dt = D0 (1‐r)t
r
D0
0,1
0,2
0,4
0,05
D5
0,1181 0,0039
D10
0,0697 0,0003
A2 qA B1 pB B2 qB A1 B2 A2 B1 A2 B2 pA qB qA pB qA qB pA1B2 pA2B1 pA2B2
D = pA1B1‐ pA pB
-D
-D
+D
coupling heterozygote A1B1/ A2B2 : 2* pA1B1 * pA2B2 = 2* ( ½ (1‐r))2
repulsion heterozygote A1B2/ A2B1 : 2* pA1B2 * pA2B1 = 2* ( ½ r)2
in equilibrium: 2* pA1B1 * pA2B2 = 2* pA1B2 * pA2B1 not in equilibrium: pA1B1 * pA2B2 ≠ pA1B2 * pA2B1 66
Lecture1.1.5
Polulation genetics
van Leeuwen
r = 0,5: unlinked loci
Hardy Weinberg equilibrium is not true when:
•Assortative (not random) mating
•Different gamete production per genotype
•Migration
•Selfing ‐ Partly selfing
•Selection
•Fixation
67
Lecture1.1.5
Polulation genetics
van Leeuwen
Assortative mating
if mated pairs are of the same genotype more often than would occur by chance
occurs with human beings, but is not related to a single gene
increases the number of homozygotes
Dissortative mating
if mated pairs are of the same genotype less often than would occur by chance
occurs in plants with self‐incompatibility systems
increases the number of heterozygotes
other examples?
Migration
migration of a fraction m
existing population: 1‐m, gene frequency: q0
new immigrants: m
gene frequency: qm
mixed population:
q1 = m qm + (1 – m) q0
= m (qm‐q0) + q0
Δq = q1 ‐ q0 = m (qm – q0 )
68
Lecture1.1.5
Polulation genetics
van Leeuwen
In plants, migration of pollen can occur. If the gene frequency is substantially different it has influence
Disequilibrium occurs, but equilibrium will be restored, if it is an unique occurance
pollen: p1 = (1‐m) p0 + m pm
q1= (1‐m) q0 + m qm
the influence of the migration depends on m and p0 ‐ pm
pollen
situation 1
migration: only pollen
genotype G0
number
frequency
p0
pm
m
0,5
0,1
0,1
0,8
0,1
0,1
0,8
0,1
0,4
p1
G0 genotypes:
A1A1
A1A2
A2A2
total
360
480
160
P
H
Q
0,36
0,48
0,16
p
0,6
q
0,4
1000
1
1
pollen cloud
q=
0,4
90000
pollen cloud from outside
qmigr. =
0,1
10000
p(pollen) = pp
q(pollen) = qp
1
p(egg cell) = pe
q(egg cell) = qe
1
mixed population: q1 = m qm + (1 – m) q0 = 69
m = ?
Lecture1.1.5
Polulation genetics
p(pollen) = pp
0,63
q(pollen) = qp
0,37
p(egg cell) = pe
0,6
q(egg cell) = qe
0,4
van Leeuwen
The generation G1 has the following genotypes:
genotype
A1A1
A1A2
A2A2
total
frequency
frequency formula
1
pp*pe
pp*qe + pe*qp
qp*qe
1
number
number G0
1000
360
480
160
After random mating: p =
Mutation
If in one genotype one gene is mutated, the chance that the gene is disappearing in next generations is very large
Usually AA Aa
Only with selfing the gene has more chance to survive in the population
u and v are mutation frequencies per generation, usely u>v if A1 is wildtype
u
Mutation rate
A1
Initial gene frequencies p0
70
v
A2
qo
Lecture1.1.5
Δq = up0 ‐ vqo
Polulation genetics
van Leeuwen
Mutation
A1A1
genotype
freq. genotypes
A1A2
A2A2
total
0,36
0,48
0,16
1
p2
2pq
q2
1
number
360
480
160
1000
p=P+ 1/2 H
0,6
q = Q + 1/2 H
0,4
frequency formula
u
0,00001
v
0,000001
p1 =
0,599994
q1 =
0,400006
at equilibrium:
pu = qv
q = u/(u+v)
Selection
Individuals can differ in viability and fertility and so contribute differently to the next generation
fitness or adaptive value or selective value
coefficient of selection is called s
contribution of ‘favoured’ genotype is: 1
contribution of genotype selected against is:
1‐s
71
Lecture1.1.5
Polulation genetics
van Leeuwen
Genotypes
A1A1
A1A2
A2A2
Initial frequencies
p2
2pq q2
Coefficient of selection
0
0
s
Fitness
1
1
gametic contributions
p2
2
q sq0
q1 0
2
1 sq0
2
1
1‐s
q2
2pq q (1 s ) p0 q0
q1 0
2
1 sq0
Total
(1‐s) 1 ‐ s q2
new gene frequency after selection
p = 1‐q
Δq = q1 – q0
sq 2 (1 q )
q
1 sq 2
Table 2.2 Falconer
p1 = 1‐q1
gene frequencie of A2 = q
Initial frequencies and
New gene
Change of
fitness of genotypes
frequency
gene frequency
A1A1
A1A2
A2A2
p2
2pq
q2
1
1 – ½ s 1-s
1
1-hs
1-s
1
1
1-s
1-s
1-s
1
q1
Δq = q1 - q
q1
q sq sq
1 sq
q1
q hspq sq 2
1 2hspq sq 2
1
2
q1
1
2
q sq 2
1 sq 2
Δq = ± ½ sq (1‐q)
Δq = ± sq2 (1‐q)
q sq sq 2
q1
1 s(1 q 2
72
2
Lecture1.1.5
Polulation genetics
van Leeuwen
Table 2.2
Falconer
gene frequencie of A2 = q
Initial frequencies
and
New gene
Change of
fitness of
genotypes
frequency
gene frequency
A1A1
A1A2
A2A2
p2
2pq
q2
1-s1
1
1-s2
q1
q1
Δq = q1 - q
q s2 q 2
1 s1 p 2 s2 q 2
approximately: no dominance: Δq = ± ½ sq (1-q)
dominance:
Δq = ± sq2 (1‐q)
sq 2 (1 q )
q
1 sq 2
if s or q is small, Δq is approximated by
approximately: q sq 2 (1 q)
dominance:
no dominance: Δq = ± ½ sq (1-q)
Effectiveness of selection depends on s and q
73
Δq = ± sq2 (1‐q)
Lecture1.1.5
Polulation genetics
van Leeuwen
no dominance
complete dominance
Generations
Change of gene frequencies, from one extreme to another.
selection of intensity s= 0,2, at different q. q2 = freq. A2A2
‐ selection against gene with frequency q
+ selection in favour of the gene with frequency q
start with heterozygous individuals
q = 0,5
aa = lethal
s = 1, h = 0
A1A1 and A1A2 equal
……………..
Fig. 2.4. Change of gene frequency under natural selection in the laboratory
74
s=1, h=0,1
A1A2 less fit
Lecture1.1.5
Polulation genetics
van Leeuwen
selection in a breeding program against aa
Genotype
Number
Frequency
After selection
Freq. after
AA
640
0,64
Aa
320
0,32
aa
40
0,04
Total
1000
1
Gene frequency A =
after selection
q1
Gene frequency a =
after selection Male
A
p0
q0
p1
q1
q0 sq0
2
1 sq0
2
a
Female
Genotype frequency:
sq 2 (1 q)
q
1 sq 2
q1
q sq 2
1 sq 2
Calculate with this formula q in following generations with selection against A2A2 = aa
2
s = ? s=1
q1
q0 q0
q0 (1 q0 )
q
0
2
(1 q0 )(1 q0 ) 1 q0
1 q0
q0 = 0,04+0,5*0,32=0,2
q1 = (0,2/(1+0,2)= 0,167
75
Lecture1.1.5
Polulation genetics
van Leeuwen
Decrease of undesired genotypes (aa) in different generations
Generation
Mass selection
1
Number of undesired
genotypes aa per
1000
40
2
28
3
20
4
16
5
13
6
10
7
8
8
7
9
6
10
5
q=
0,2
0,167
0,020
0,125
0,016
0,111
0,012
0,100
0,010
0,091
0,008
0,083
0,007
0,077
0,006
0,071
0,005
Removal of undesired gene after flowering:
After Selection:
male side
640 320 40 1000
PM
qM
Female side
Freq.
♀
0,028
0,143
Genotype: AA Aa aa total
Number: 640 320 40 1000
♂
q2 =
pF =
qF = = 0,8
= 0,64 + ½ *0,32
= ½ *0,32 + 0,04
= 0,2 640 320 ‐‐
0,67 0,33
960
1
0,64 + ½ *0,32 = 0,833
½ *0,32
= 0,167
76
Lecture1.1.5
Polulation genetics
van Leeuwen
Genotype frequency is:
Male
A
0,8
a
0,2
Female
A
0,833
0,666
0,167
a
0,167
0,134
0,033
The genotypes in the next generation are:
AA Aa aa total
666 301 33 1000
With selection after flowering, removal of aa is even slower
Small populations
random drift
with a small population by coincidence a genotype or gene may disappear
differentiation between sub‐populations
leads to genetic differentiation between subpopulations
uniformity within subpopulations
individuals become more and more alike in genotype
homozygosity increases
revealing of recessive genes results in inbreeding depression
77
Lecture1.1.5
Polulation genetics
78
van Leeuwen
Lecture 1.2.1
Reproductive Systems
van Leeuwen
1.2.1 Reproductive systems: pollination, reproduction and plant breeding
‐ Brief overview of gametogenesis and seed formation
‐ Pollination mechanisms
‐ Functional vs. Morphological‐Structural Flower Biology
‐ Definition of sex types in flowering plants
‐ Terminology and classification
Common features among life cycles 1. Two phases (n, 2n): relative importance varies
2. Mitosis: geometric expansion of tissue
3. Meiosis:
numerical and genetic reduction, recombination
4. Fertilization
79
Lecture 1.2.1
Reproductive Systems
Male: Development of pollen grains
A a microspores
A
B b c van Leeuwen
C
1. meiosis
A
C microspore mother cell or mycrosporocyte
b
b
C
a
n
n
B
c n
n
a
4 functional
microspores
each microspore will develop in ripe pollen grain
2. nucleus in pollen grain divides in two: tube nucleus (for tube growing) and generative (sperm) nucleus
microspore mother cell
( binucleate pollen) 3. Sperm nucleus divides in two
(trinucleate pollen)
80
B
c Lecture 1.2.1
Reproductive Systems
Pollen grains with nucleus germinating pollen grain
Gametogenesis of Angiosperms (Flowering Plants
1. Microsporogenesis 2. The two haploid nuclei of a pollen grain have different destiny
• one haploid nucleus becomes generative nucleus and later on divides into two haploid sperm nuclei (3). • The other haploid nucleus of pollen grain remains undivided and is called tube nucleus.
4. Now the pollen grain is ready for pollination. male development
81
van Leeuwen
Lecture 1.2.1
Reproductive Systems
Female: Development of egg cell and central cell/polar nuclei
1. Megasporogenesis • The sporogenesis which occurs in the female part of the flower, the ovary to produce female reproductive cells, the megaspores or embryo sacs, is called megasporogenesis. van Leeuwen
n
2n
meiosis
n
n
n
megaspore mother cell or megasporocyte • The diploid cells of ovary which undergoes the meiotic division to form megaspores, are called megaspore mother cells or megasporocytes • A single megasporocyte of the ovary undergoes the meiosis to form four haploid megaspores, all of which get linearly arranged.
1 functional
megaspore
• Only one megaspore survives Female development a. Out of these four megaspores, three megaspores degenerate and the remaining megaspore undergoes three successive mitotic divisions without any intervening cytokinesis to form a large cell with eight haploid nuclei.
b. This octanucleate cell is the immature embryo sac. The embryo sac is surrounded by maternal tissues of the ovary called integument and also by the megasporangium or nucellas. It has a minute opening called micropyle at its one end for the penetration of pollen tube in it. 82
Lecture 1.2.1
Reproductive Systems
van Leeuwen
MMC = megaspore mother cell Nu= nucellus V =vacuole
OIn= outer and inner (IIn) integuments
FM = functional megaspore seven cells: two synergid cells (SC); one egg cell (EC); one central cell (CC) carrying two polar nuclei (PN); and three antipodal cells (AC)
CNN = two polar nuclei have fused to form the central cell nucleus
Female development c. The eight nuclei of embryo sac have different destiny. Out of eight nuclei, three nuclei migrate towards micropyler end and two of them (called synergids) degenerate.
d. The remaining third nucleus develops into an egg nucleus. e. Another group of three nuclei migrates towards opposite end of the embryo sac and are called antipodals. The antipodals also degenerate soon. f. The remaining two nuclei are called polar nuclei. They remain in the centre of the embryo sac and both unite to form a single diploid fusion nucleus. g. The embryo sac is now mature and ready for fertilization.
83
Lecture 1.2.1
Reproductive Systems
van Leeuwen
The events of "double fertilization" of the egg and polar nuclei by the two sperm cells
• The process of pollination being accomplished, the pollen tube grows through the stigma and style toward the ovules in the ovary. • The germ cell in the pollen grain divides and releases two sperm cells which move down the pollen tube. The events of "double fertilization”
• Once the tip of the tube reaches the micropyle end of the embryo sac, the tube grows through into the embryo sac through one of the synergids
which flank the egg. – One sperm cell fuses with the egg, producing the zygote which will later develop into the next‐generation embryo. – The second sperm fuses with the two polar bodies located in the center of the sac, producing the nutritive triploid endosperm tissue that will provide energy for the embryo's growth and development
84
Lecture 1.2.1
Reproductive Systems
Maize
85
van Leeuwen
Lecture 1.2.1
Reproductive Systems
van Leeuwen
In the ovule the macrospore developes to the embryo sac
macrospore (after meiosis) has 3 mitotic divisions,
8 nuclei
1st division
4 nuclei
2 nuclei
3st division
86
2nd division
Lecture 1.2.1
Reproductive Systems
van Leeuwen
embryo sac with 8 nuclei
egg
Pollen tube growth and fertilisation
recognition by secretion of species specific proteins at stigma
stigma
style
ovary
Higashiyama et al. (2003) Curr. Opin. Plant Biol. 6: 36-41.
pollen tube is attracted by nuclei in ovule
MA Johnson and D Preuss (2002)
Developmental Cell 2: 273-281.
87
Lecture 1.2.1
Reproductive Systems
van Leeuwen
normal fertilization
Hamer et al., 1990
Pathway of pollen tube growth through the female sexual tissues of a flower. (B) Stigma and upper part of the style. Pollen grains (p) on the stigma surface where they germinate to produce pollen tubes, which grow through the central transmitting tract (tt) of the style. (C) Transmitting tract tissue consists of files of elongated cells, which are separated at maturity by a secreted mucila (m). The pollen tubes grow in the mucila between files of cells. The sperm cells (sc) are contained within the tips of the pollen tubes. The mucila contains mixtures of proteins, glycoprorins, and protoglycans as nutrients for the growing pollen tube
(D and E) Micrographs showing pollen tubes within the style of N. alata after a compatible pollination, stained with a fluorochrome and viewed by fluorescent microscopy. (D) Section including stigma. Pollen grains and tube walls fluoresce. (E) Section within the style adjoining that shown in (D). 88
Lecture 1.2.1
Reproductive Systems
van Leeuwen
Embryo
future seed skin
After fertilisation zygotes developes into embryo by mitotic division
Seed contains:
Embryo (2n) +
Seed coat (2n)
Endosperm (3n) + +
Pollination, reproduction and plant breeding
• Pollination mechanisms consist of:
• Sexual differentiation
• Microsporogenesis, • Macro(mega)sporogenesis
• Structural differentiation of flowers
• Pollen dispersal ecology
• Structural or physiological barriers and facilities to fertilization
• Each of the above has implication on plant breeding and crop production
89
Lecture 1.2.1
Reproductive Systems
van Leeuwen
Pollination mechanism and breeding of new cultivars
• Three main phases in a plant breeding program in all crops:
– Generate genetic variation in form of a population
– Select and develop elite genotypes
– Synthesise the elite genotypes into a cultivar
• Programs are cyclical in nature, only the new cultivars break out of the cycle requiring
– Maintenance
– Multiplication
– Distribution
Generalization goes only this far, pollination mechanisms dictate: • Breeding methods – will spend lots of time on this.
• Type of cultivars (OP, hybrids, clones), • Manner in which the cultivars are – Maintained (outcrossing rates and purity requirements)
– Multiplied – extremely important in hybrid crops (hand vs. insect pollination)
• Distributed (seed, tubers, seedless fruits, cut flowers)
90
Lecture 1.2.1
Reproductive Systems
van Leeuwen
Morphological‐Structural Flower Biology
• Darwin and even before
• Characterised with elaborated description of morphology
Functional Flower Biology – last 30 years
• Dynamic functional approach to flower biology
– Interrelation between pollen transfer and flower structure
– Flower induction
– Photoperiodic and thermoperiodic influences
– Environmental and hormonal influences
– Genetic sex determination
– Natural cross‐pollination rates
– Physiology of self‐incompatibility
– Pollination ecology etc...
Induction of Flowering – Photoperiod
Many plants are induced to flower only when they receive photoperiods either longer or shorter than a critical length. Short‐day plants (SFP) require long nights, while long‐day plants (LDP) require short nights in order to flower. Exposure to light for only a few minutes in the night can prevent some short‐day plants from flowering. This response is controlled by phytochrome proteins. Bewley et al. 2000. In Buchanan, Gruissem and Jones, eds. Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists.
UCDAVIS
91
Lecture 1.2.1
Reproductive Systems
van Leeuwen
Induction of Flowering – Temperature
In addition to light, temperature is another environmental signal used by many plants to trigger flowering. Many plants, especially biennials, require a period of chilling (temperatures below 10 C) to induce flowering. This is termed vernalization. Plants often must reach a minimum size before they are capable of being vernalized, although there are exceptions to this, as for wheat or lettuce, which can be vernalized during germination. Winter wheat and many common vegetables (onions, beets, celery, carrots, most Brassicas) require vernalization, while in others, chilling only speeds flowering (lettuce, spinach). The latter is known as facultative vernalization. UCDAVIS
Floral Meristems Develop Inward from the Outer Whorl
UCDAVIS
92
Lecture 1.2.1
Reproductive Systems
van Leeuwen
The ‘ABC’ Model of Flower Organogenesis
According to the “ABC” model, flower differentiation is controlled by three types of regulatory genes.
Genes
B A Whorl 1 Organs
A A & B B & C C (A & C repress each other)
C 2 3 4
Organ
M. Yanofsky and R.J. Schmidt (1999) Trends in Plant Sci. (poster insert)
UCDAVIS
Definition of sex types in individual flowers
• Hermaphrodite –both stamens and carpel(s)
• Staminate (or androecious) – only stamens, no carpels
• Pistillate – (or gynoecious, carpillate) – only carpel(s), no stamens
Definition of sex types in flowering plants
e.g. in Cucurbitaceae
Plants with only female flowers
= gynoecious
Plants with only male flowers
= androecious
Plants with female and male flowers
= monoecious
Plants with bisexual flowers
= hermaphrodite
Plants with male and bisexual flowers
= andromonoecious
93
Lecture 1.2.1
male flower
male
androecious
Reproductive Systems
bisexual flower
female flower
monoecious
van Leeuwen
female
gynoecious
andro‐
monoecious
hermaphrodite
dioecious: seperate male and female plants
tomato grass wheat
cucumber squash maize spinach
potato, etc.
hermaphrodite or bisexual flower unisexual flower
asperagus
cucumber squash maize spinach
monoecious plant
94
dioecious plant
Lecture 1.2.1
Reproductive Systems
van Leeuwen
Method of reproduction
‐
asexual reproduction
from a part of the plant a new plant grows (clone)
‐
sexual reproduction
seed is combination of egg cell and pollen grain
* cross fertilization
* self fertilization
range from 100% self pollination – partly self pollination – 100% cross pollination
Self fertilization enhanced by flower structure (autogamy):
Stigma and pollen grain of one flower are ripe at the same time
Cleistogamous: Flowers are closed untill the pollination is ready (lettuce, bean, pea, wheat) •Pistil and stigma grow through a ‘tube’ of stamens, while growing through they are pollinated (lettuce, tomato, pepper)
95
Lecture 1.2.1
Reproductive Systems
van Leeuwen
Cross fertilization with bisexual flowers
Stigma and pollen grain are in the same flower not ripe at the same
time (leek, carrot, onion) protogynous: first stigma is ripe than pollen
protandrous: first pollen is ripe than stigma (onion)
Selfincompatibility: e.g. with cabbage egg cell is genetically incompatible with pollen grain of the same plant
Flower structure:
Stamen much shorter than stigma
Pistil and stigma grow through a ‘tube’ of stamens, while growing through the stigma is closed or not ripe (chicory)
Cross fertilization by Insects:
Attractive flowers for colour, nectar , smell
Sticky pollen grains
Room in the flowers for insects
Flowering over a longer time span
96
Lecture 1.2.1
Reproductive Systems
van Leeuwen
Wind:
Light and small pollen grains
Plants produce a lot of pollen grains (cloud of grains)
Pollen grains can easily be taken by the wind
Stigma with wide surface
Asexual forms of reproduction
• Asexual propagules outside the floral region
– Natural or artificial propagules
• Clonal maintenance of self‐incompatible varieties (avocado, some grasses)
• Fixation of heterozygocity and hybrid vigour (roses, potato)
• Multiplication of material under conditions in which plants do not flower (sweet potato)
– Types of propagules
•
•
•
•
•
Stem, leaf, root cuttings
Layering (rooting the stem)
bulbs, tubers
Grafting (joining of parts of plants together)
Meristem culture (pathogen free material)
97
Lecture 1.2.1
Reproductive Systems
van Leeuwen
Asexual propagules within the floral region: Apomixis
Modes of apomictic reproduction
•
Agamospermy – parthenogenetic seed (parthenos = virgin)
– Gynogenesis – female haploid gametophyte → new haploid spofophyte
• Pollen can play a role to activate, but no fertilization
• Pseudogamy: pollination is absolute requirement although male and female gametes never unite – provides the necessary stimulation
– Androgenesis – male haploid gametophyte → new haploid spofophyte
– Apospory ‐ development of an embryo of a flowering plant outside the embryo sac, from a cell of the nucellas (megasporangium) or chalaza
• Most common mode of apomixis (Kentucky Bluegrass)
– Diplospry – formation of embryo from e.g. unreduced megaspore (2n gametophyte = 2n sporophyte)
– Adventitious Embryony – and embryo arises from the diploid nucleus tissue surroungind the embryo sac (common in Citrus)
• Vivipary (from floral axils and branches)
– Vegetative proliferation (bulbs etc...)
– Female somatic tissue → diploid sporophyte
Importance of apomictic seeds
• Uniformity in seed propagation (many)
• True breeding of F1 hybrids (Poa) • Removal of viruses from the clones of vegetatively
propagated plants
• Distribution of modes of reproduction among cultivated plants
– See summary pages 17‐28, in Frankel & Galun paper (in handouts)
98
Lecture 1.2.1
Reproductive Systems
van Leeuwen
Reproduction in the Angiosperms ‐ Consequences for Breeding Plant Breeding Methods Allogamy
Population (heterogeneous mixture of heterozygous individuals) Plant 1: Aa Bb cc Dd EE Ff Plant 2: aa Bb Cc dd EE Ff Plant 3: AA bb Cc Dd Ee Ff Inbred (homogeneous mixture of homozygous individuals) Plant 1: AA bb cc DD EE ff Plant 2: AA bb cc DD EE ff Plant 3: AA bb cc DD EE ff Hybrid (homogeneous mixture of heterozygous individuals) Plant 1: Aa bb Cc Dd Ee Ff Plant 2: Aa bb Cc Dd Ee Ff Plant 3: Aa bb Cc Dd Ee Ff Reproduction in the Angiosperms ‐ Consequences for Breeding Plant Breeding Methods Autogamy
Old land race (heterogeneous mixture of homozygous individuals) Plant 1: AA bb cc DD EE ff Plant 2: aa BB CC dd ee FF Plant 3: AA bb CC DD ee ff Cultivar (homogeneous mixture of homozygous individuals) Plant 1: AA bb cc DD EE ff Plant 2: AA bb cc DD EE ff Plant 3: AA bb cc DD EE ff Apomixis / Clone (homogeneous mixture of heterozygous individuals) Plant 1: Aa Bb cc Dd EE Ff Plant 1: Aa Bb cc Dd EE Ff Plant 1: Aa Bb cc Dd EE Ff
99
Lecture 1.2.2
Pollination Control
Pollination control
•Mechanical control
•Chemical control
•Genetic control
Mechanical control of pollination
Bisexual flowers and unisexual flowers on monoecious plants:
to prohibit self pollination
prohibiting self pollination:
•hand emasculation: laborious, chance for mistakes
•mechanical detasseling (maize)
•water spraying on pollen to kill the pollen (lettuce)
No 100% security that selfing is prohibited (control!)
100
van Leeuwen
Lecture 1.2.2
Pollination Control
Mechanical control of pollination
to avoid unwanted pollination from neighbour plants
•covering plants or flowers
•closing big flowers with clip (squash)
crossing between two grass plants
squash
chemical control of pollination
•chemical hybridizing agents (CHA)
•gametocides
•male sterilants
for those crops where hybrid seed production may be interesting, but with no availability of male sterility:
big expectations, but not very succesful
101
van Leeuwen
Lecture 1.2.2
Pollination Control
van Leeuwen
Only known to be used in China and India for developing of rice hybrids
disadvantages:
unsafe for human beings
effectiveness is highly stage‐specific and genotype‐specific
induced male sterility is not 100% secure
advantages:
only 2 lines needed for a hybrid (3 in case of male sterility)
not dependent on the existance of a male sterility system in a crop
hybrid seed production with Chemical Control Agent
source: IRRI
102
Lecture 1.2.2
Pollination Control
van Leeuwen
Genetic control:
•unisexual flowers
•male sterility
•self incompatibility
sex expression:
Maize
Cucurbitaceae
Spinach
Asparagus
monoecius
dioecius/monoecius
dioecius/monoecius
dioecius
Selfing not impossible with monoecius plants
Spinach:
female and male
cucumber: female flower
Asparagus
female and male
103
Lecture 1.2.2
Pollination Control
van Leeuwen
Male sterility
can be defined as a condition in which a normally bisexual flower is not able to produce viable, fertilising pollen
True male sterility:
•stamen are absent
•stamen are not producing pollen •the pollen grain is unviable
•or cannot germinate and fertilize normally to set seeds
Functional male sterility:
Anthers fail to release their contents even though the pollen is fertile
Male sterility
Three systems:
•Genetic (nuclear, genic) male sterility (GMS)
•Cytoplasmic male sterility (CMS)
•Cytoplasmic‐genetic male sterility (CGMS)
No selfing of mother lines:
necessary to develop maintainer lines
104
Lecture 1.2.2
Pollination Control
van Leeuwen
Genotypic male sterility
Mendelian inheritance due to nuclear not cytoplasmic genes 1) Arisen as spontaneous mutants in most cases:
‐ high frequency of mutation. 2) Identified in 175 species. ‐ ms (recessive‐‐most) or Ms (dominant‐‐few) Recessive: single genes ~70%; multigenes ~15% (monocot) and 23% (dicot)
Dominant mutants: 7% (dicot) and 15% (monocot)
Many nonallelic genes are known in some species (e.g. 60+ in maize, tomato 55, etc.)
Kaul, 1988
Genetic male sterility (GMS)
Seed production
Is used in tomato and peppers One recessive gene
mm = male sterile Mm = male fertile MM = male fertile
male sterile flower
fertile flower
A‐line (mother line)
mm
C‐line (father line)
x
MM
Hybrid seed
(Mm)
105
Lecture 1.2.2
Pollination Control
van Leeuwen
Genetic male sterility (GMS)
Maintenance of parent lines
mm =
Mm =
MM =
male sterile
male fertile
male fertile
A-line
(mother line)
mm
x
B-line
(maintainer line
Mm
C-line
(father line)
MM
crossing
mm
mother line
+
self ing
Mm
MM
maintainer line
father line
maintainer line is isogenic to mother line
Functional or positional male sterility:
viable pollen grains are available, but the anthers don’t release the pollen
• Non‐opening of the anthers
• Pollen not released but viable • Hand pollination possible
106
tomato:
ps ‐2 mutant
Lecture 1.2.2
Pollination Control
van Leeuwen
Figure 1: Microscopic observation of anthers of ps‐2ABL and wild type (WT; Moneymaker). Tomato
wild type
Benoit Gorguet, Phd thesis WUR, 2007
wild type
fertilization is possible manually
ps‐2 mutant
107
ps‐2 mutant
Lecture 1.2.2
Pollination Control
van Leeuwen
Environment‐sensitive genic male sterility
Male sterility is controlled by nuclear gene expression, which is influenced by environmental factors such as temperature, daylength (EGMS)
Is reported in several crops
used only in rice for hybrid seed production
advantage: monogenic or oligogenic: easy to transfer
maintenance by selfing is possible (in other environment)
disadvantage : male sterilty can be less than 100% due to environmental conditions
Cytoplasmic male sterility (CMS)
Male sterility caused by variation in mitochondrial DNA (in cytoplasma)
The phenomenon is regarded as a mitochondrial ‘mutation’
or ‘deficiency’ causing the normal developmental program
of male gamete production to fail (Budar et al. 2003)
cytoplasma is usually only transfered via the egg cell
(not via the pollen grain)
maternal inheritance
(S): male sterile plasma
(N) or (F): normal or fertile plasma
108
Lecture 1.2.2
Pollination Control
van Leeuwen
Cytoplasmic male sterility (CMS)
male fertile plant male sterile plant (N) (S) cytoplasma nucleus (2n) (N) (S) nucleus (n) nucleus (n) egg cell pollen grain egg cell Cytoplasmic male sterility (CMS)
Seed production
motherline (S) X pollinator/father (N) So hybrid is male sterile
hybrid (S) 109
Lecture 1.2.2
Pollination Control
van Leeuwen
maize
carrot
onion
sugar beet
rice
cabbage
rape seed
sun flower
male sterile flower
Cytoplasmic genic male sterility (CGMS)
the male sterility can be restored by restorer genes (Rf)
restorer genes are genes in the nucleus which restore the fertility of the male sterile plasma
idiotype = cytoplasmatype + genotype
most restorer genes are dominant
110
Lecture 1.2.2
(S) rf rf = Pollination Control
male sterile
van Leeuwen
(S)
rf rf
(S) Rf rf =
(S) Rf Rf
male fertile
(N) Rf Rf =
(N) Rf rf
(N) rf rf
male fertile
(S)
Rf Rf/rf
(N)
rf rf
(N)
Rf Rf/rf
Cytoplasmatic male sterility (CMS) with restorer genes
Genes are necessary to make hybrid male fertile (for seed or fruit production) These genes in maintainer lines can also restore male fertility in mother lines !
Ster‐/Fert rapeseed
111
Lecture 1.2.2
Pollination Control
van Leeuwen
Cytoplasmatic male sterility (CMS)
•is found in nature
•can also be caused by an interspecific hybrid (natural or not)
Cytoplasmatic male sterility (CMS)
is caused by bad cooperation of plasma and nucleus
Fusion of cytoplasma with the nucleus of another species is called a cybrid
examples: chicory CMS from sunflower via protoplast fusion
Brassica‐CMS from Raphanus via protoplast fusion
112
Lecture 1.2.2
Pollination Control
cybride
Self Incompatibility
Self incompatibility = inability to a fertile, hermaphrodite seed plant to produce zygotes after self‐pollination (de Nettancourt, 1977)
one of the methods to encourage outbreeding
Self‐incompatibility is a mechanism for self‐recognition that results in rejection of self pollen by the female somatic tissues (Haring et al., 1990)
Self‐ Incompatibility: A Self‐ Recognition System in Plants
Haring et al., 1990
Science, New Series, Vol. 250, No. 4983 (Nov. 16, 1990), pp. 937‐941 113
van Leeuwen
Lecture 1.2.2
Pollination Control
van Leeuwen
Most Incompatibility systems are controlled by a single locus, the S locus, with multiple alleles. highly polymorphic locus more than 40 S alleles in natural populations
Self incompatibility is developmentally regulated and the SI barrier can often be overcome by hand‐pollination of immature flowers with mature pollen from the same plant (in this way, homozygous plants can be produced)
SI is overcome or weakened by increased temperature
or other stress conditions
gametophytic SI
the incompatibility reaction is initiated by the interaction of a product of the haploid genome of the male gametophyte (carried within the pollen grain) and a product of the diploid
genome of the female tissue of the sporophyte, the pistil. If the pollen carries the same allele as one of the two in the pistil, fertilization is not achieved
S1
no fertilization
S1 S2
114
Lecture 1.2.2
Pollination Control
van Leeuwen
sporophytic SI
the incompatibility reaction is between factors carried by the pollen but specified by the diploid tissues of the pollen parent and a product of the female pistil. In the simpliest case, pollen tube growth is arrested if either one or two of the alleles of the pollen parent is also present in the pistil.
S1
male = S1 S2
S2
no fertilization
S1 S3
gametophytic systems:
pollen tube growth is arrested within the style and involves contact between the pollen tube and the mucilage secreted by cells of the transmitting tract inhibition in style
Solanaceae, Liliaceae, Rosaceae
sporophytic systems:
tube growth is arrested at the stigma surface or soon after penetration and involves contact between the pollen grain or emerging pollen tube and material secreted into the cell walls or onto the surface of the stigmatic papillae
inhibition on stigma
Brassicaceae, Asteraceae, Poaceae
115
Lecture 1.2.2
Pollination Control
van Leeuwen
gametophytic incompatibility S3
S1 S2
S1
S4
fertilization
S2
S1
S2
S1 S2
S1
S1 S3
no fertilization
S2
S2 S4
gametophytic incompatibility S3
S1
S1 S2
S1
partial fertilization
S2
S1
S2
S1 S2
S1
S1 S3
116
S2
no fertilization
Lecture 1.2.2
Pollination Control
van Leeuwen
gametophytic incompatibility fertilization
S1 S2 x S3 S4 => S1 S3 + S1 S4 + S2 S3 + S2 S4
partial fertilization
S1 S2 x S1 S3 => no fertilization
S1 S2 x S1 S2 => S1 S3 + S2 S3
no seed set
Sporophytic incompatibility
Incompatibility genes : S alleles
Incompatibility if :
the father plant has at least one allele in common with the mother plant
S1
father = S1 S2
S2
S1 S3
117
no fertilization
Lecture 1.2.2
Pollination Control
van Leeuwen
Sporophytic incompatibility
S4 S
3
S3
S4
S1 S2
fertilization
S1
S2
S1 S4
S2
S1 S2
no fertilization
S2 S3
S2
S1
S1
S1
S1 S3
S2
S2 S4
Sporophytic incompatibility
S3
S1
S1 S2
S1
no fertilization
S2
S1
S2
S1 S2
S1
118
S2
no fertilization
Lecture 1.2.2
Pollination Control
Sporophytic incompatibility with codominance
fertilization
no fertilization
no fertilization
S1 S2 x S3 S4 => S1 S3 + S1 S4 + S2 S3 + S2 S4
S1 S2 x S1 S3 => no seed set
S1 S2 x S1 S2 => no seed set
partial fertilization is not posible
Alleles can interact S1 S2 :
•Codominance: S1 and S2 react both
•dominance: S1 dominates S2 (S1 > S2)
•dominance: S2 dominates S1 (S2 > S1)
Can be different in father and mother!!!
119
van Leeuwen
Lecture 1.2.2
Pollination Control
van Leeuwen
Sporophytic incompatibility with codominance in mother and dominance in father
S1
S2
S1 S3
fertilization
S1 S2
S2 > S1
S1 S3
S1
father
=> S2 S2
mother
=> S1 S3
S3
S1 S1
S2 S3
Incompatibility system makes selfing difficult
selfing is possible by:
•bud pollination by hand young buds are opend, anthers removed
stigma is pollinated with ripe pollen of other flower of the same plant
•CO2 in closed greenhouse
120
Lecture 1.2.2
Pollination Control
van Leeuwen
Not all S‐alleles are effective, also weak S‐alleles exist
Effectivity of S‐alleles is dependent or environmental conditions
Influence of developmental stage of flower bud on the seed setting (Kakizaki)
% seed settin
60
50
40
30
20
10
0
-6
-4
-2
Selfing AA
Zelfbest.
Selfing BB
Zelfbest.
Crossing AxB
Kruising
AxB
0
2
4
6
developmental stage of flower bud in
days
Sporophytic Incompatibility is used in hybrid seed production in Brassica oleracea
hybrid seed production maintaining parent lines parent line 1 parent line 2 parent line 1 parent line 2 S1S1 X S2S2 S1S1 S2S2 ... crossing insects bud pollination or CO2 Hybrid parent line 1 parent line 2 S1S2 S1S1 S2S2 121
Lecture 1.2.2
Pollination Control
van Leeuwen
Incompatibility causes selfing and crossing barriers in many species
for example:
•in wild, outcrossing tomato species
•in grasses
•in fruit species (Rosaceae, apple and prune)
•in outcrossing Asteraceae (e.g. chicory)
is pollinated by
pollination scheme for Dutch apple varieties x = compatible
x = self‐compatible
122
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
1.2.5. Basic statistics
Statistics is the science, pure and applied, of creating, developing, and applying techniques by which the uncertainty of inductive inferences may be evaluated
(Steel et al., 1997)
plant breeding:
‐ creating variation by bringing together several genetic resources
‐ select the best genotypes at certain stages in the breeding process
start breeding process: many different genotypes, one to a few plants per genotype
in selection process: decreasing number of different genotypes, increasing number of plants per genotype
end: 1 genotype with many plants introduction of new variety
(or 1 group of narrowly related genotypes
123
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
General procedure
•breeding purposes
•genetic resources
•to recombine
parents with special requirements
wide variation, all different genotypes
•to select
•to finish off the variety
selection procedure depends on variety type
one new variety, least possible variation within the variety, uniformity
selection decisions based on observations of individual plants or a group of plants
how to select the best genotype?
124
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
Variation sources
Genetics 1. Characteristics are determined by one to three genes (Mendel, segregation)
2. Characteristics are determined by more than three genes (Mendel, no visable segregation, continuous variation)
Environment Characteristics are influenced by the environment (mostly polygenic characteristics)
80
%
60
•resistent or susceptible
40
•red or white flower
20
0
1
2
3
4
5
6
7
8
9
•bitter or not bitter
resistentie
resistance
Qualitative characteristic = •trait, which shows a clear phenotype
•segregation
•discontinuous variation
•usually no influence of the environment
125
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
aantal
10
X
9
X
8
X
X
7
X
X
X
6
5
4
3
2
1
X
X
X
X
X
X
X
X
X
X
X
X
X
X
X
X
X
X
observations plants in table or histogram
X
X
X
X
X
X
X
X
X
X
X
X
X
X
X
X
X
X
20
21
22
23
24
25
26
27
28
29
lengte van de maïskolf in cm
length of cob in maize in cm
43 plants, length of cob in maize in cm
%
25
23
21
25
24
28
22
25
26
25
21
29
22
25
26
24
28
27
23
23
26
23
23
24
24
40
35
30
25
20
15
10
5
0
27
27
26
24
25
20
24
24
26
26
22
26
25
24
25
25
27
25
Quantitative characteristic/trait
1
2
3
4
5
6
7
8
9
Traits:
resistentie
resistance
numbers
•No distinct segregation
length
•Difficult to divide in groups
weight
•Continuous variation
days after sowing
•Usually influence of the environment
content
•Based on many genes
percentage infected leaves
•Numeric
scores
126
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
polygenic trait in wheat (pure line variety) variation by genotype or environment?
polygeen, veel milieu-invloed
X mean
100
number of aantal plante
plants
80
60
Reeks1
40
20
110
107
104
101
98
95
92
89
86
83
80
77
74
71
68
0
length in cm
average = 93.4 cm
This image cannot currently be display ed.
1 gene, little influence of environment
aa
Aa
AA
25
15
aa
10
Aa
AA
5
75
73
71
69
67
65
63
61
59
57
55
53
51
49
47
0
45
number of plan
20
length in cm
1 gene, a little influence of environment
25
20
15
Series1
10
5
0
0
20
40
length in cm
127
60
80
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
1 gene, more influence of environment
25
aa Aa AA
20
aa
Series1
Aa
Series2
15
Series3
AA
Series4
all
10
5
67
65
63
61
59
57
55
53
51
49
47
45
0
length
1 gene, intermediate inheritance
number of plan
25
20
aa
Reeks1
15
10
Aa
Reeks3
5
AA
0
0
20
40
60
80
Reeks2
Reeks4
all
length
1 gene, strong influence of environment
aa Aa AA
25
20
aa
Series1
Aa
Series2
15
Series3
AA
Series4
all
10
5
67
65
63
61
59
57
55
53
51
49
47
45
0
length
1 gene, strong influence of environment
25
20
aa Series1
Series2
AaA
Series3
Aall
Series4
15
10
5
0
0
20
40
length
128
60
80
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
number of
plants
value
1
45
6
50
15
55
20
60
15
65
6
70
1
75
3 genes, intermediate inheritance, determine 1 trait
AaBbCc selfing:
F2: segregation (see table)
you see hardly any segregation
3 loci, intermediate
25
number
20
15
Reeks1
10
5
0
0
10
20
30
40
length in cm
129
50
60
70
80
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
Phenotype = genotype + environment
Plant 1
2 3 4 5
In order to select the best genotype:
1. decrease the variable influence of the environment
2. use statistical analysis to cope with all unidentified variable effects
quantitative trait
Sugar content in sugar beets
traits that show variation are called variables
Y = variable
Y1 = first observation of variable
Y2 = second observation of variable
130
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
presentation of set of observations as a vector
21
Y1
Y=
Y2
Yn
=
24
29
or is presented as a row vector:
Y’ = { Y1, Y2, ……….. Yn } = { 21, 24,……….. 29 } in the classification of a variable, we have ideas about the relative frequencies of a variable
we observe probabilities of the occurence of a certain value
sugar content in sugar beet:
an observation of 18% is as expected, but 50% is not
131
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
probability density function for a discrete, qualitative trait:
probability of getting a baby girl is 0.5 probability of getting a baby boy is 0.5
only two possible values
probability distribution function for a continuous, quantitative trait:
probability of getting a baby with a weight of 3 kg is 0.4 probability of getting a baby with a weight of 5 kg is 0.01
all values possible within a certain range with different probabilities per value
Are the data collected all data of a population or only the data of a sample?
population: consists of all possible values of a variable
weight of all newborn babies in the Netherlands in 2009
sample: consists of all possible values of a part of the population
weight of 100 newborn babies (at random) per province in the Netherlands in 2009 (totally 1200 babies)
132
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
the data of a sample, usually are used to make an inference about the population
population: good definition
weight of all newborn babies in the Netherlands in 2009: are babies included, who are born too early (definition of too early?)
sample: has to be representative
weight of 100 newborn babies (at random) per province : number of inhabitants per province is different, representative?
Experimental data soybean plants can be presented in table or histogram
Frequency
Histogram yield data
3
7
8
5
13
7
18
18
23
32
28
41
33
37
38
25
43
22
48
19
53
6
58
6
63
3
68
1
45
40
35
number of plan
yield
30
25
Reeks2
20
15
10
5
0
3
8 13 18 23 28 33 38 43 48 53 58 63 68
Yield in grams
133
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
Experimental data soybean plants can be presented in table or histogram
yield
Frequency
3
7
8
5
13
7
18
18
23
32
28
41
33
37
38
25
43
22
48
19
53
6
58
6
63
3
68
1
population:
all plants of the soybean variety
population is characterized by quantities called parameters, like the arithmetic mean
sample: number of randomly chosen plants from population
values are observed in experiment
sample is characterized by quantities called statistics, like the arithmetic mean
Experimental data soybean plants can be presented in table or histogram
yield
Frequency
3
7
8
5
13
7
18
18
23
32
28
41
33
37
38
25
43
22
48
19
53
6
58
6
63
3
68
1
number
229
mean
31.9
population is characterised by the arithmetic mean = μ
sum of values of all items
μ=
number of items
sample is characterised by the arithmetic mean = Y
Y1 + Y2 + ……….. Y229
Y =
229
134
Lecture 1.2.5
yield
Basic Statistical Concepts
van Leeuwen
Experimental data soybean plants
Frequency
3
7
8
5
13
7
18
18
23
32
28
41
33
37
38
25
43
22
48
19
53
6
58
6
63
3
68
1
number
229
mean
31.9
sample is characterised by the arithmetic mean =
Y
Y =
n
i 1
Yi
n
or
Y
i
i
n
Y1 + Y2 + ……….. Y229
229
= 31.9
Frequency distribution of yield data
number of plan
50
40
30
Reeks1
20
Mean is the same for both experiments
10
0
0
20
40
60
80
yield in grams
Dispersion is different:
Frequency
lower graph has less dispersion
number of plan
60
50
40
30
Reeks1
20
10
Variance is measure of dispersion
0
0
10
20
30
40
50
60
70
yield in grams
Y = 31.9 in both cases
135
Lecture 1.2.5
Basic Statistical Concepts
Yi
van Leeuwen
variance of a population:
99
100
101
2
Y
i
2
i
N
103
104
variance of a sample:
102
105
s
102
sum
816
Y
102
2
Y
i
Y
2
i
n 1
Sum of Squares = SS
Yi
Yi Y Yi Y 2
Y
i
Y
2
i
variance of a sample:
99
var(Y ) s
100
2
Y
i
n 1
101
103
Y
2
i
n‐1: degrees of freedom (df)
104
n‐1 is used in a sample, because one degree of freedom is used in estimating Y
102
105
102
SSY i Y i Y
2
sum
816
Y
102
var=
n‐1 = var = standard deviation = s = √s2 = SD = √(varY) =
136
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
2
Yi
Yi
99
9801
100
10000
101
10201
103
10609
104
10816
102
10404
105
11025
102
10404
sum
816
83260
Y
102
SS
Y
Y
i
_
2
i
i
Y
2
i
Y
i
=
2
i
n
= 83260 – (816)2/8 = 28 n = 8
s2 = 28/7 = 4
83232 =8162/8
Y
i
i
2
= C (correction factor)
n
Standard error of the mean
If we take 4 samples of a population
we can calculate the mean of each sample
means are less variable than single observations
the estimated mean of the population will be ‘the mean of the means’
we can calculate the variance of the mean
Y
2
population
2
n
sY
2
s2
n
sample
SE sY
s2
n
standard error of the mean (SE)
137
Lecture 1.2.5
Basic Statistical Concepts
coefficient of variability:
can be used to compare experiments
CV
100 * s
%
Y
what is regular in your crop?
Normal distribution
bell‐shaped curve symmetric about the mean
with high numbers of observations the histogram can be refined so much that it is a smooth curve
138
van Leeuwen
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
Normal distribution
•bell‐shaped curve symmetric about the mean
•normal distribution is described by μ and σ
•higher σ means more dispersion of data
Probability distribution:
P (Y < μ ) = 0.5
number of plants
culm length in spring wheat
120
100
80
60
40
20
0
60
70
80
90
100
110
length in cm
skew distribution
mean 83.4
a lot of statistical analyses assume a normal distribution, sometimes unjustified
median = 81.5 (50%)
modus = 80 (value of Y for which the number is at highest
139
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
probability that
Y > 85 = 0,006210
http://davidmlane.com/hyperstat/z_table.html
normal distribution: at large numbers and quantitative traits
standard normal distribution:
95% of the individuals of a population are between ‐2 and +2 140
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
So in a standard normal distribution 99,73% of the values are between ‐3 and + 3
So in a standard normal distribution 68,27% o
the values are between ‐1 and + 1
µ = 0, σ = 1
In a normal distribution 68,27% of the values are between ‐1 σ and + 1σ
µ = σ = 141
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
1. Normal distributions are symmetric around their mean.
2. The area under the normal curve is equal to 1.0.
3. Normal distributions are denser in the center and less dense in the tails.
4. Normal distributions are defined by two parameters, the mean (μ)
and the standard deviation (σ).
5. 68% of the area of a normal distribution is within one standard
deviation of the mean.
6. Approximately 95% of the area of a normal distribution is within
two standard deviations of the mean.
standard normal distribution
f(Z)
0,45
0,4
0,35
0,3
0,25
0,2
0,15
0,1
0,05
0
-4
-3
-2
-1
0
1
Z
P( Z z )
Table A.4
142
2
3
4
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
standard normal distribution N(0,1)
P(Z 1) 0.8413
P(Z 1.15)
0.8749
Probability that a random value will be < 1.15
function, which describes the normal distribution N(μ,σ)
From standard normal curve N (0,1) to a normal curve with μ and σ2 :
Z
Y
normal distribution
0,5
f(Y))
0,4
0,3
0,2
0,1
0
-4
‐4σ
‐3σ
-3
-2
‐2σ
-1
‐1σ
μ0
Y
143
1
1σ
2σ2
3
3σ
4
4σ
Lecture 1.2.5
Z
Basic Statistical Concepts
van Leeuwen
Y
P(Z 1.15)
P(
Y
1.15) 0.8749
If μ = 10 and σ = 2
P(
Y 10
1.15)
2
P (Y 12,3)
0.8749
http://davidmlane.com/hyperstat/z_table.html
P (Y 12,3) 0,8749
144
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
P( Z 2) P( Z 2) P( Z 2)
1 ‐ 0.9773 = 0.0227
P(2 Z 2)
1 – 2 * 0.0227 = 0.9546
P( 2 Y 2 ) 0.9546
So roughly:
μ ± 2σ covers 95% of the values of Y
N(
normal distribution
1,2
1
0,8
0,6
0,4
0,2
0
16
16,5
17
17,5
18
sugar content in beets
Y
s
s
sinterval
(95% of plants has a sugar content of 17.35 + 0.66
between 16.69 and 18.01)
145
18,5
Lecture 1.2.5
Basic Statistical Concepts
van Leeuwen
If we want to make an estimation of the mean of a population, we also want to know how reliable that estimation is: can we be confident that the estimation is a good representation of the reality.
verdeling knollengte
tuber length in potato
aantal knollen
no. of tubers 8
n = 29
7
6
5
4
3
2
1
0
Y = 58
Reeks1
var(Y)=3,6
50
52
54
56
58
60
62
64
66
68
70
knollengte in mm
tuberlength in mm
aantal knolle
no. of tubers
n = 29
Y = 58
verdeling knollengte
tuber length in potato 8
6
Reeks1
4
2
0
var(Y)=10,6
50
52
54
56
58
60
62
64
66
68
70
knollengte in mm
tuber length in mm
If we want to make an estimation of the mean of a population, we also want to know how reliable that estimation is: can we be confident that the estimation is a good representation of the reality?
aantal knolle
no. of tubers
verdeling knollengte
tuber length in potato
8
n = 29
6
Reeks1
4
2
var(Y)=10,6
0
50
52
54
56
58
60
62
64
66
68
Y = 58
70
knollengte in mm
tuberlength in mm
aantal knolle
no. of tubers
verdeling knollengte
tuber length in potato
25
n = 130
20
15
Reeks1
10
5
Y = 58
var(Y)=7,6
0
50
52
54
56
58
60
62
64
knollengte in mm
tuberlength in mm
146
66
68
70
Lecture 1.2.5
Basic Statistical Concepts
95% of the observations is between:
Y – 2s < Yi < Y+ 2s
n = 29
Y = 58
95%: < Yi < 95%: < Yi < 95%: < Yi < var(Y)=3,6
s = 1,90
n = 29
Y = 58
var(Y)=10,6
s = 3,25
n = 130
Y = 58
var(Y)=7,6
s = 2,76
Reliable estimation of the mean
standard deviation of μ = standard error
SE = / n
95% confidence interval of the mean
Y- 2 SE < Y + 2 SE
with a probability of 95% μ is between
Y 2 SE and Y 2SE
147
van Leeuwen
Lecture 1.2.5
Basic Statistical Concepts
95% probability that μ is between: Y 2 SE Y 2 SE
n = 29
Y = 58
95%: < µ < 95%: < µ < 95%: < µ < s = 1,90
s/√n = 0,35
n = 29
Y = 58
s = 3,25
s/√n = 0,6
n = 130
Y = 58
s = 2,76
s/√n = 0,24
148
van Leeuwen
Lecture 1.3.1
Cytogenetics
1.1.4. Cytogenetics
Cytogenetics is a branch of genetics that is concerned with the study of the structure and function of the cell, especially the chromosomes
Some slides from
David Stelly
Texas A&M University
History
1665 Hooke (England) –
described the cell
17th century: discovery of microscope
19th century: use of dyes for staining intracellular structures (nucleus and others) observation of nuclear division, 149
van Leeuwen
Lecture 1.3.1
History
Cytogenetics
plant cell
botany
1665 Hooke (England) –
described the cell
cell membrane
cell nucleus
17th century: of microscope
discovery van Leeuwen
mitochondria
cytoplasma
chloroplasts
cell wall
19th century: use of dyes for staining intracellular structures (nucleus and others) observation of nuclear division, botany
Linnaeus
1753 Linnaeus –
published “Species Plantarum”; binomial nomenclature of plant taxonomy
150
Lecture 1.3.1
Cytogenetics
van Leeuwen
History Koelreuter, Joseph Gottlieb 1733‐1806 Koelreuter was a plant hybridizer. Between 1760 and 1766: first series of systematic experiments in plant hybridization (Nicotiana paniculata x N. rustica). Hybrid offspring resembles both parents
Thus for the first time it was found that the pollen grain has an important part in determining the characters of the offspring. Mendel 1865
Versuche über Pflanzen‐
Hybriden (1865)
von Gregor Mendel.
http://www.mendelweb.org/MWpaptoc.html
151
Lecture 1.3.1
Cytogenetics
van Leeuwen
Cell biology
Each cell in the plant contains all genes
In each tissue different genes are expressed/switched on
Each genome contains about 30.000 genes
Some genes are homozygous and essential for the growth and development of the plant
Some of the genes have 2 alleles
different genotypes within a species
some genes are multi‐allelic
not all genes are active in each cell switch (promotor) for each gene (differentation)
Cell Division
Prokaryotes
Eukaryotes (plants, animals, fungi)
(bacteria)
BINARY FISSION
chromosome
chromosome segregation
Somatic cells
diploid
MITOSIS
Germ cells
haploid
MEIOSIS
Karyokinesis
Partition of the nucleus
Cytokinesis
Division of the cytoplasm & cell membrane
152
Lecture 1.3.1
Cytogenetics
van Leeuwen
Common features among life cycles 1. Two phases (n, 2n): relative importance varies
2. Mitosis: geometric expansion of tissue
3. Meiosis:
numerical and genetic reduction, recombination
4. Fertilization
Common features among most eukaryotic organisms • Nucleus
contains the chromosomes (except mitosis & meiosis), aids protection, function, regulation …
• Chromosomes
100 um
~2 m of DNA per cell
packages DNA & proteins for mitosis, meiosis; other functions, e.g., expression regulation
• Mitosis (and cytokinesis)
copies and distributes DNA & certain proteins for somatic growth
• Meiosis (+/‐ cytokinesis)
copies, recombines, reduces chromosome number and distributes DNA … for sexual reproduction
153
Lecture 1.3.1
Cytogenetics
van Leeuwen
“Genome” (nuclear)
Monoploid
Diploid
Unreplicated
Double
helix
Replicated
Basic Chromosome Structure
Unreplicated
Telomere
Centromere
(dynamic composition)
Replicated
Semi‐conservative
DNA Synthesis
(using both strands
as templates)
Telomere
(one chromatid)
(one LONG double helix of DNA, plus associated proteins and RNAs, per unreplicated chromosome)
2 “sister chromatids” per replicated chromosome
154
(two LONG double helices of DNA, plus associated proteins and RNAs)
Lecture 1.3.1
Cytogenetics
van Leeuwen
Chromosomes have different forms
All have a centromere which plays a role in doubling and dividing the chromosomes
Centromere
sister chromatides
Doubled chromosome
Mitosis ‐‐> Genetic Fidelity
Unreplicated
g
Replicated
Semi‐conservative
DNA g
g
Synthesis
(using both strands
as templates)
“Sister
Chromatids”
g
g
g
g
Mitosis: “sister chromatid disjunction”
155
Lecture 1.3.1
Cytogenetics
van Leeuwen
2 homologous chromosome 1
2 homologous chromosome 2
doubled chromosome with two chromatids
nucleic membrane disappears
2 homologous chromosome 1
2 homologous chromosome 1
156
doubled chromosome 1
seperated chromatides
Lecture 1.3.1
Cytogenetics
van Leeuwen
mitosis
A a B b A a c B b c C C diploid cell, 2n=6
AaBbCc
A a c
no recombination
C S = synthesis‐phase = doubling of chromosomes
Gap2 = normal cell activities
M = mitosis‐phase nucleus and cell divide
Gap1 = normal cell activities
157
Cell cyclus
B b Lecture 1.3.1
Cytogenetics
van Leeuwen
Mitosis
• Extremely high fidelity (chr & genes)
• EXCEPTIONS:
– Developmentally planned deviations
– Induced, chemicals, pathogenic, symbiotic
– Mistakes (factors: G, T, E, G*E, other)
Root‐tip
(“section” cut from a paraffin‐embedded tip, using a “microtome”)
• RAMIFICATIONS:
– Genome dosage: polyploidy, – Genetic reduction: somatic recombination (rare), somatic chromosome substitution
– Chromosomal aberations, aneuploidy and segmental aneuploidy: misdivision, translocation, fragmentation, and nondisjunction…, genome instability.
Generalized Life Cycle ‐‐ DNA Content
1 C
• Unlike mitosis: meiosis is divided into two rounds of division (one DNA replication, but two divisions)
158
Lecture 1.3.1
Cytogenetics
van Leeuwen
2 homologous chromosome 1
2 homologous chromosome 2
doubled chromosome with two chromatids
paired, doubled chromosomes
159
Lecture 1.3.1
Cytogenetics
van Leeuwen
Terminology
Homologous chromosome pair
(“homologs”, “homologues”)
g
g G
Homologous chromosome pair
G
H
H h
h
NON‐homologous chromosomes
(“non‐homologs”)
2 “Sister
chromatids”
2 “NON‐sister
chromatids”
160
Lecture 1.3.1
Cytogenetics
van Leeuwen
Meiosis ‐‐> Balanced, Genetic Infidelity
“Homologous chromosome pair
g
g G
“Homologous chromosome pair
G
H
[1] Independent assortment of genes in non‐homologous
chromosome pairs.
H h
h
MEIOSIS VS MITOSIS
•
Mitosis maintains genetic fidelity, whereas meiosis destroys it. •
Mitosis maintains chromosome number, whereas meiosis halves it. Expected meiotic products:
1/2 H 1/2 h .
1/2 G GH : Gh : gH : gh
1/2 g 1/4 : 1/4 : 1/4 : 1/4
diploid cell, 2n=6
meiosis
AaBbCc
Abc
A a meiosis
B b aBC
c C A c
b A a
b c
B C 161
a
B C Lecture 1.3.1
Cytogenetics
van Leeuwen
diploid cell, 2n=6
meiosis
AaBbCc
AbC
A a meiosis
B b aBc
c C A A b C
A a a
b C
A a B b a
c c A a B b B B A a B b B b c c c c C C C C meiosis
2 ABC and
2abc
or
2 ABc and
2abC
or
2 Abc and
2aBC
or
2 AbC and
2aBc
AaBbCc
1/8 ABC + 1/8 abc + 1/8 ABc + 1/8 abC + 1/8 Abc + 1/8 aBC + 1/8 AbC + 1/8 aBc
meiosis gives variation by new combinations of chromosomes and genes
162
Lecture 1.3.1
Cytogenetics
van Leeuwen
Meiosis ‐‐> Homologous recombination (“crossing over”)
“Homologous chromosome pair
G
G g
g
B
B
b
b
CHROMATIDS:
Each homologous recombination event (crossover) involves just 2 of the 4 chromatids, so there are just 50% recombinant products. Chiasmata are a cytological manifestation of crossing over.
[2] Homologous recombination or crossing over (CO) of genes in non‐sister chromatids.
Sister chromatids do not recombine. WT gametes (parental): Rec gametes (nonparental): 1:1, 1:1, GB : gb
Gb : gB
Meiosis ‐‐> Balanced, Genetic Infidelity
“Homologous chromosome pair
G
G g
maternal
g
paternal
Keep in mind: each homologous pair includes 1 maternally derived homolog (from both maternal chromosomes), and 1 paternally derived homolog, .
163
Lecture 1.3.1
Cytogenetics
van Leeuwen
diploid cell, 2n=6
Crossing over
AaBbCc
A a ABCD ABCd
meiosis
B b abcD abcd
c D d
C A C
B D
A C
a
B
d b D
c a
b c d
meiosis causes genetic variation by:
1. different combination of chromosomes (and genes)
2. new combinations of genes by crossing over
164
Lecture 1.3.1
Cytogenetics
van Leeuwen
The Most Important Consequences of Meiosis
1.
Balanced reduction • chromosome number, e.g., 4 ‐‐> 2
• gene content, e.g., Aa ‐‐> A or a
2.
Recombination of genes
• Between non‐homologues: Independant Assortment (more chromosomes more recombination)
• Between homologues: recombination, limited by linkage Changes in Chromosome Number
Ploidy
Monoploid
Ploidy ‐ Levels & Types
Diploid
Related diploid
Polysomic polyploid
Tetrasomic tetraploid
Disomic polyploid
Disomic tetraploid
165
Lecture 1.3.1
Cytogenetics
van Leeuwen
Terminology Disomic Polyploids (“allopolyploids”)
Homologous chromosome pair
Homologous chromosome pair
(“homologs”, “homologues”)
g1
g1 G1
G1
g2
g2 G2
G2
Homeologous chromosomes
(a.k.a. homoeologous chromosomes)
Similar genetic content, but usually meiotically independent (usually no synaptic pairing or homologous recombination).
Haploids ‐ Fertility?
Diploid
WHY?
Sterile monoploid
Tetrasomic tetraploid
(+ or ‐ fertility)
Fertile diploid
Disomic polyploid
(fertile)
Sterile diploid
WHY?
166
Lecture 1.3.1
Cytogenetics
Aneuploidy:
incomplete set of chromosomes
one cromosome more or less
aberrant phenotype
monosomic: 2n‐1
nullisomic: 2n‐2
trisomic 2n+1
wheat 2n=6x=42
phenotype, if certain chromosome is lacking
167
van Leeuwen
Lecture 1.3.1
Cytogenetics
van Leeuwen
“Alien Additions” ‐‐
various wheat alien addition lines with chromosomes from Elymus, a related genus.
E
W F1
Disomic additions (d, g, h, I): WW + various (II)E
Ditelosomic addition lines (e, j): WW + various (ii)E Monotelosomic addition (f): WW + iE
Normal gamete development
first division second division
normal meiosis 4 haploid gametes
168
Lecture 1.3.1
Cytogenetics
van Leeuwen
unreduced gametes
2n plant 2n gametes
first division restitution
(FDR‐gametes)
second division restitution
(SDR‐gametes)
if this happens to the whole genome 2n gameten
if this only happens to 1 chromosome pair
aneuploid gametes
169
Lecture 1.5.1
Mean Separation T-Test
Mean separation, t‐test
1. Testing of hypotheses
2. Brief review of:
‐Standard normal distribution, Z value, standardization
3. Distribution of sample means
4. t‐distribution
5. t‐test
‐Population mean and a specific value
‐Two or more means
‐Two independent samples with equal variances
‐Paired observation
testing of hypotheses
hypothesis:
statement that something is true
•null hypothesis = H0
•alternative hypothesis = H1 (it is not true)
rejection of null hypothesis:
based on the value of a testing parameter (t)
reject H0 if |t| > (critical value from table)
significant result
170
van Leeuwen
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
Statistical decision True state of null hypothesis
H0 = true
H0 = false
Accept H0
Correct
Type II error
Reject H0
Type I error
Correct
Statistical decision is:
based on the value of a testing parameter (t)
reject H0 if |t| > (critical value from table) significant result
testing parameter (t) = calculated with observations Yi
table based on distribution of test parameter
Decision can be correct or incorrect (=error)
Decisions and their outcomes
Statistical decision
Acceptance region
(i.e. non‐
significant)
Decision is to:
Accept H0
Data are from a population with:
H0 = true, H1 = false H0 = false, H1 = true
Correct decision
Probability should be high
Incorrect decision
Type II error
Symbol: 1 – α = confidence coefficient
Probability should be low
Symbol: β
Reject H1
Rejection region
(i.e. significant)
Correct decision
Incorrect decision Probability should Type I error
be high
Reject H0
Probability should be low
Symbol: α = significance level
Accept H1
Significance level mostly 0.01 ‐ 0.05
171
Symbol: 1 – β = power
Lecture 1.5.1
Mean Separation T-Test
Statistical decision True state of null hypothesis
H0 = true
H0 = false
Accept H0
Correct
Type II error
Reject H0
Type I error
Correct
The type I error we want very low,
is called significance level or reliability level :
= 0.05 or 0.01,
is the propability that we make a type I error
Type II error means that we don’t reject H0, while it is false; is called discriminating power
Normal Distribution
Standard Normal Distribution: mean µ=0; std σ = 1, standardization by Z
Y
172
van Leeuwen
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
• Assuming that a population of the percent sucrose of sugar beets is normally distributed with a mean of 12.2% and a standard deviation equal to 2.26%.
• What is the probability of a given sugar beet having a sucrose concentration greater than 10% and less than 14%? • We can state the problem Y
mathematically keeping in mind: Z
P(z<Z) = P(z< (10‐12,2)/2,26) =
P(z< ‐0,97) = P(z>0,97 = 0,166
P(z>Z) = P(z> (14‐12,2)/2,26) =
P(z>0,8) = 0,212 Geng, Table A‐4.
P(10% < sucr. % < 14%) = 1 – 0,212 – 0,166 = 0,622
• This is equivalent to finding the probability that Z is between ‐0,97 and 0,80
• (iii) Find the area (1 ‐ A ‐ B) which is equal to ( 1 ‐ 0.2119 ‐
0.1660) = 0.62. • The probability is 62% that a randomly drawn sugar beet from this population will have a sucrose content between 10% and 14%. B=0,166
‐0,97 0
Z
173
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
• What is the probability of a randomly drawn sugar beet from this population having sugar concentration:
• >14%
• >15%
• <10% Distribution of sample means:
‐ when a population is sampled (e.g. replicated plots), it is customary to calculate the statistics (e.g. means)
‐ therefore generating a population of sample means with a mean and variance on its own.
‐ Assumption:
‐ The samples drawn are approximately normally distributed
‐ The statistics computed are close approximation to the original population.
‐ In a population of means made of n observation, Y 2
Y
the mean and equals
Y
and
Y 2
2
n
174
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
Distribution of sample mean differences: ‐ Consider two large populations from which samples were drawn with samples of sizes or n1 and n2. The means and variances are:
‐ An additional approximately normal distribution is generated by taking the differences between all possible means d = Y‐X.
‐ The parameters are: μd and σ2d
‐ When:
Square root of variance of mean differences is standard error of difference between sample means
Student t‐distribution
‐ Already established that a population of sample means is approximately normally distributed.
2
Y and Y 2
n
‐ This population can be standardized to become a standard normal distribution.
‐
the actual is rarely known and if it is replaced by an estimator ; ‐ the distribution is no longer normal, but similar called t‐distribution
175
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
The value of t is calculated as
t
Y Y
sY
s2
df = n‐1
n
There is a t‐distribution for every sample size (n‐1 = df), distributions approaching normal with increase of df. Exercise
‐
t ≥ t0 ; t ≤ t0 ; t ≥ t0 ≤ t;
tdf;α;tail = critical value, dependant on:
df: degrees of freedom = n‐1
α: significance level
tail: two tailed or one tailed t
tdf;α;tail
test parameter tcalc. =
t
Y Y
sY
s2
one tailed table
n
P(tcalc. > tdf;α;tail)
example:
n=10 df=9
α = 0,05
one tailed
tdf;α;tail = 1,833
176
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
Example
• Distribution of t (df = 5 – 1 = 4) compared to Z. • The t distribution is symmetric and somewhat flatter than the Z, lying under it at the center and above it in the tails. • The increase in the t value relative to Z is the price we pay for being uncertain about the population variance
Students t‐test can be used for:
t‐test
‐Population mean and a specific value
‐Two or more means
‐Two independent samples with equal variances
‐Paired observation
177
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
Testing hypothesis that a population mean is a specific value
• μ and σ2 = mean and the variance of a population
• a random sample of size n is drawn from the population and the sample mean and the variance, and s2 and are computed. hypothesis, the test criterion:
• To test the H0 : Example: compare mean with fixed value µ0
Machine to fill sacs with beans has been set at 1000 g H0 : µ0 = 1000 H1 : µ0 ≠ 1000 Sample is taken to check if the setting is right
Statistical decision True state of null hypothesis
H0 = true filling weight = 1000g
H0 = false
filling weight ≠1000g
Accept H0
Correct
(filling weight = 1000) 95% probability Type II error
=power = β Reject H0
Type I error
Significant level = α = 5%
Correct (filling weight ≠ 1000)
178
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
filling machine: 1000 g per sac, μ0 = 1000
sac
1
2
3
4
5
6
7
8
9
10
sum
mean
weight Yi Y
982
1003
973
961
997
979
991
1009
988
969
9852
985
2
H0: filling weight is 1000
testing parameter (t): upon which the decision is taken
derived from sample data Y and s
1. how much is the deviation from the mean?
2. how high is the variance?
testing parameter (t):
t
Y 0
s/ n
filling machine: 1000 g per sac, μ0 = 1000
sac
1
2
3
4
5
6
7
8
9
10
sum
mean
2
weight Yi Y
982
9
1003
324
973
144
961
576
997
144
979
36
991
36
1009
576
988
9
969
256
9852
2110
985
234,44
s2
15,31
s
4,84 s/√n
H0: filling weight is 1000
testing parameter (t): upon which the decision is taken
1. how much is the deviation from the mean?
2. how high is the variance?
Y μ0) / (s/√n) =
t = ( ‐
179
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
filling machine: 1000 g per sac, μ0 = 1000
t
H0: filling weight = 1000
Y 0
s/ n
Decision rule: reject H0 if |t| > 2.26
if ‐2.26 < t < 2.26 critical value
than with P = 95%, H0 is correct
tdf;α;tail = t9;0,05;2tailed =
in this case: t = ‐3.09 and |t| > 2.26
H0 is rejected: machine sets are not correct
α = 0,05
area = 0,025
‐tdf
0
rejection region acceptance region Y 0
t
s/ n
area = 0,025
area = 0,95
tdf
rejection region
Students t‐distribution with n‐1 degrees of freedom
180
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
Decision rule: reject H0 if |t| > 2.26
Where can you find the ‘critical value’?
α = 0,05
area = 0,025
area = 0,025
area = 0,95
tdf;α;tail
‐tdf
degrees of freedom:
0
tdf
rejection region acceptance region rejection region
n‐1 = 9
table A.6:
DF = 9
α = 0,05 2‐tailed=
0,025 1‐tailed
One tailed table : P(tcalc. > tdf;α;1 tail) tdf;α,tail = t9;0,05;2 =2.26
If we only want to know if > μ
Y
Y
0, than H1: > μ
0
Decision rule: reject H0 if t > 1.83
α = 0,05
tdf;α;tail =t9;0,05;1 = 1.83
area = 0,95
0
acceptance region t
area = 0,05
(from table)
tdf
rejection region
One tailed table : P(tcalc. > tdf;α;1 tail) Y 0
s/ n
Students t‐distribution with n‐1 degrees of freedom
181
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
The mean of the sample is significant different from 1000 g
or
confidence interval of the mean:
Y t*
s
n
= 985 ± 2.26 * 4.84 Y
974 < < 996
does not contain the value of 1000
new sample
sac
1
2
3
4
5
6
7
8
9
10
sum
mean
weight
982
1007
980
955
997
979
900
1100
988
970
9858
985
Yi Y ) 2
9
484
25
900
144
36
7225
13225
9
225
22282
2475,78
s2
49,76
s
15,73 s/√n
H0: filling weight is 1000 g
t
Y 0
s/ n
‐2.26 < t9;0,05;2 < 2.26 with P=95%
conclusion?
182
Lecture 1.5.1
Mean Separation T-Test
Symbol
Description
P‐value
~
indication for significance
0.05<P<0.10
*
significant
0.01<P<0.05
**
highly significant
0.001<P<0.01
***
very highly significant
P<0.001
van Leeuwen
Test for two or more means
• t ‐ statistic can be used to test hypotheses about the mean of a population of mean‐differences.
• Consider two populations with means μ1and μ2. – A random sample is drawn from each population to test the null hypothesis that e.g. μ1= μ2 . t is defined by:
The calculation of the standard deviation term depends on whether:
• Two populations have a common variance σ2
• Two samples are of equal size
• The observation are paired
183
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
How to decide if differences between varieties are significant?
30
25
aantal
20
Reeks1
15
Reeks2
10
5
40
41
.5
37
38
.5
34
35
.5
31
32
.5
28
29
.5
26
.5
25
0
aantal
lengte komkommer
N2 = 200
x1 = 29
x2 = 38,5
s1 = 1.4
s2 = 1.4
Reeks1
Reeks2
21
.5
23
.5
25
.5
27
.5
29
.5
31
.5
33
.5
35
.5
37
.5
39
.5
41
.5
43
.5
45
.5
N1 = 200
18
16
14
12
10
8
6
4
2
0
lengte komkommer
N1 = 200
N2 = 200
x1 = 29
x2 = 38,5
s1 = 2.6
s2 = 2.6
How to decide if differences between varieties are significant?
6
5
aantal
4
Reeks1
3
Reeks2
2
1
45
43
41
39
37
35
33
31
29
27
25
23
21
0
lengte komkommer
6
N1 = 40
N2 = 40
x1 = 29
x2 = 38,5
s1 = 3.3
s2 = 3.3
5
4
Series
2
3
2
1
0
21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39
N1 = 40
N2 = 40
x1 = 29
x2 = 31
s1 = 3.2
s2 = 3.2
184
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
Two independent samples with equal variances
Example Treatment Replications e.g. tonnes / hectare
A
38.8
37.6
37.4
35.8
B
40.9
39.2
39.5
38.6
38.4
39.8
Total
188
198
Mean
37.6
39.6
The question to be answered: is the mean difference, 39.6‐37.6 =2.0, really due to the treatments or to a chance occurrence?
H0: d 1 2 0
H1: d 1 2 0
From the observations we calculate:
d Y1 Y2
2
s1 and s2
2
Variance of the difference of the mean = var + var
(Y1 )
(Y2 )
sd
2
2
2
2
s
s
1 2
n1 n2
SED sd
2
s1 s2
n1 n2
Two independent samples with equal variances
Example Treatment Replications e.g. tonnes / hectare
A
38.8
37.6
37.4
35.8
B
40.9
39.2
39.5
38.6
38.4
39.8
Total
188
198
Mean
37.6
39.6
The question to be answered: is the mean difference, 39.6‐37.6 =2.0, really due to the treatments or to a chance occurrence?
test parameter t =
t
d d
2
SED sd
sd
2
s1 s2
n1 n2
standard error of the mean
with df = n1 + n2 ‐ 2
If equal sample size (n), the best estimate of the common variance is pooled variance):
2
2
s 2 ( s1 s2 ) / 2
SED sd
185
2s 2
n
with df = 2(n‐1)
Lecture 1.5.1
Mean Separation T-Test
van Leeuwen
Two independent samples with equal variances
Example Treatment
A=1
B=2
Replications e.g. tonnes / hecatar
38.8
37.6
37.4
40.9
39.2
39.5
35.8
38.6
Total
188
198
38.4
39.8
Mean
37.6
39.6
The question to be answered: is the mean difference, 39.6 ‐ 37.6 = 2.0, really due to the treatments or to a chance occurrence?
i.
Determine the mean for each sample
d Y1 Y2
ii.
Determine the variance of each sample
s1 and s2
iii.
Determine the standard error of the mean difference
SED sd
iv.
Compute the t‐statistics
2
sum
mean
var=s2
d
sd2
SED=sd
t
A
B
38,8 40,9
37,6 39,2
37,4 39,5
35,8 38,6
38,4 39,8
188
198
37,6 39,6
1,34 0,725
2
0,413
0,64
3,112
2
t
With df = (n1‐1)+(n2‐1) = 8
t
d d
sd
2
2
s1 s2
n1 n2
d d
sd
2
3,112
0,64
df = 8
α = 0,05
tail: 2 tailed
critical value:
tdf;α;tail = t8;0,05;2tail = 2,306
1 tailed table
t = 3,112 > t8;0,05;2tail = 2,306 H0 is rejected
The difference between treatment A and B is significant and caused by the treatment and not by coincidence with a probability of 95%
186
Lecture 1.5.1
Mean Separation T-Test
student t ‐ distribution table
testing parameter
t
d d
van Leeuwen
df = degrees of freedom = n1 + n2 ‐ 2
sd
LSD = least significant difference:
difference, which is just significant
LSD t df ; ;tail * sd
If the observed difference > LSD d
H0 is rejected; the difference is significant
at α = 0,05 and larger n tdf;α ≈ 2
Paired Observation
• If the pairs of observations are correlated this test will increase the ability to detect differences
• The variance of differences (paired) should be smaller than the variance of individuals. • The df = number of pairs – 1, therefore the variance must be less than 2 to compensate for the reduced df due to pairing. Treatment
Replications e.g. tonnes / hectare
A=1
B=2
B‐A
38,8
40,9
2,1
d j Y2 j Y1 j
d j d
37,6
39,2
1,6
37,4
39,5
2,1
35,8
38,6
2,8
38,4
39,8
1,4
Total
Mean
188
198
10
37,6
39,6
2
2
sd var(d )
2
2
sd sd / n
sd sd / n
187
t
d d
sd
Lecture 1.5.1
sum
mean
d
Mean Separation T-Test
A
B
38,8 40,9
37,6 39,2
37,4 39,5
35,8 38,6
38,4 39,8
188 198
37,6 39,6
2
sd2
sd2/n
√(sd2/n)
t
van Leeuwen
d Y1 Y2 d j 2
dj=B‐A
2,1
1,6
2,1
2,8
1,4
10
2
2
0,295
0,132
0,36
5,506
sd ( d j d ) 2 ) /(n 1) 0,295
2
2
sd
t
sd
0,36
n
d d
sd
2
5,5
0,36
df=n‐1=4
critical value:
tdf;α;tail = t4;0,05;2tail = 2,571
t = 5,5 > t4;0,05;2tail = 2,57 H0 is rejected
The paired difference between treatment A and B is significant and caused by the treatment and not by coincidence with a probability of 95%
Summary of t‐test
‐ Samples are used to estimate the means and variance of populations
‐ These estimates generate a family of distributions (for each sample size) that are similar to normal distribution and are called (Student) t‐distributions.
‐ Mean comparisons are done using the t‐test
‐ t – term and pooled variance are calculated based on the type of comparison and hypothesis
188
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
van Leeuwen
1.5.2. Analysis of Variance (ANOVA)
plotnr.
variety
yield
2
A
41
4
A
42
8
A
47
yield test with 3 varieties 12
A
50
completely random design
1
B
45
5
B
49
9
B
55
10
B
55
3
C
40
6
C
41
7
C
45
11
C
42
552
46
189
12 observations (plots)
randomized
B
C
A
A
C
B
A
B
A
C
C
B
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
yield test with 3 varieties completely random design
12 observations (plots)
It is possible to analyse all combinations of 2 varieties with a t ‐ test
With the F ‐ test it is possible to analyse first if there is a difference between varieties anyway
Experiment with treatments i
and observations j per treatment
t = number of treatments
r = number of replicates
Yij = ith treatment and jth observation
Yi. = mean of ith treatment (with j observations)
Y.. = mean of i treatments with j observations
190
van Leeuwen
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
hypothesis:
statement that something is true
•null hypothesis = H0
•alternative hypothesis = H1 (it is not true)
rejection of null hypothesis:
based on the value of a testing parameter reject H0 if F > (critical value from table)
significant result
Statistical decision True state of null hypothesis
H0 = true
H0 = false
Accept H0
Correct
Type II error
Reject H0
Type I error
Correct
The type I error we want very low,
is called significance level or reliability level :
= 0.05 or 0.01,
is the propability that we make a type I error
Type II error means that we don’t reject H0, while it is false
is called discriminating power
191
van Leeuwen
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
H0 :
van Leeuwen
Y1. Y2. Y3.
H1: there are differences between varieties
with the F tests variances are compared: is the variance caused by variety differences larger than the variance by coincidence/error
F =
variance by variety differences variance by coincidence/error
> 1
or > critical value
F = test parameter
variety test with onions, yield in kg/plot
Yij = + var.eff
Y
between + var.effwithin =
..
Y.. + var.effi + plot.effj
error
variety effect = ─ Yi.
Y..
Yij ─ error effect = Yi.
observation variety A, plot 1 = mean of the total experiment + variety effect (A) + error(1) (coincidence effect within a variety)
192
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
van Leeuwen
observation variety A, plot 1 = mean of the total experiment + variety effect + error
Yij = + var.eff
Y..
i + errorj
var(Y) = var(between varieties) + var(error)
We want to compare the variance due to the variety effect with the variance due to error
F =
variance by variety differences variance by coincidence/error
> critical value
to calculate SS values, for all observations effects have to be seperated and squared
Sum of Squares = SS
SStotal = SSbetween varieties + SSerror
MStotal = SStotal / dftotal
(comparable with the general calculation of the variance)
variation within varieties = variation by coincidence = standard error = remaining variation
193
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
van Leeuwen
dfbetween var. = number of varieties ‐ 1 = 2
dferror = number of observations ‐ number of varieties =12‐3=9 dftotal = number of observations ‐ 1 = 12‐1 = 11
Mean Square is calculating the variance
MSbetween var = SSbetween var /dftbetween var
MSerror = SSerror / dferror
plot
variety
yield
yield per
effect
per plot
variety
variety
2
A
41
45
4
A
42
45
8
A
47
45
12
A
50
45
1
B
45
51
5
B
49
51
9
B
55
51
10
B
55
51
3
C
40
42
6
C
41
42
7
C
45
42
11
C
42
42
sum
552
552
mean
46
194
‐1,0
Y1. (41 42 47 50) / 4 45
variety effect = Yi. Y..
= 45 – 46 = ‐1
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
plot
variety
yield
yield per
effect
per plot
variety
variety
2
A
41
45
4
A
42
45
8
A
47
45
12
A
50
45
1
B
45
51
5
B
49
51
9
B
55
51
10
B
55
51
3
C
40
42
6
C
41
42
7
C
45
42
11
C
42
42
sum
552
552
mean
46
yield
variety per plot
plot
SSvariety
‐1,0
1
0
168
yield per
effect
effect
variety
variety
error
2
A
41
45
‐1
4
A
42
45
‐1
8
A
47
45
‐1
12
A
50
45
‐1
1
B
45
51
5
5
B
49
51
5
9
B
55
51
5
10
B
55
51
5
3
C
40
42
‐4
6
C
41
42
‐4
7
C
45
42
‐4
11
C
42
42
‐4
sum
552
552
0
mean
46
195
van Leeuwen
‐4
error effect =
Yij ─ Yi.
= 41 – 45 = ‐4
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
yield
variety per plot
plot
yield per
effect
effect
variety
variety
error
SSerror
van Leeuwen
SSvariety
1
16
2
A
41
45
‐1
4
A
42
45
‐1
1
8
A
47
45
‐1
1
12
A
50
45
‐1
1
1
B
45
51
5
25
5
B
49
51
5
25
9
B
55
51
5
25
10
B
55
51
5
25
3
C
40
42
‐4
16
6
C
41
42
‐4
16
7
C
45
42
‐4
16
11
C
42
42
‐4
16
sum
552
552
0
mean
46
‐4
140
168
If variety effect is 0, no difference is expected between the both variances:
reject H0 if MSbetween var / MSerror > 1 (at a large number of observations)
We look at the critical value at the F‐table:
F‐value = MSbetween var / MSwithin var > critical value (H0 is rejected)
(at less observations)
196
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
t = number of treatments
r = number of replicates
ANOVA table
Source of variation
treatment
error
total
SS
SST
SSE
SStotal
MS
F
df
t‐1 SST/(t‐1) MST/MSE
t(r‐1) SSE/(t(r‐1))
rt‐1
critical value of F from table A.6 at P = 0,05
dependant on dfT and dfE
or calculate P via Excel (dependant on F and dfT and dfE SSvariety SSerror
168
SStotal
140
308
df
SS
van Leeuwen
df
MS
F
variety
error
total
Critical value with P = 0,05 (from F‐table A.6) = F
Conclusion? (with Excel calculated: P = 0.029
197
critical value P=0.05
P
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
van Leeuwen
plot
variety
yield
1
B
45
variety
2
A
41
A (1)
45
‐1
3
C
40
B (2)
51
5
4
A
42
C (3)
42
‐4
5
B
49
6
C
41
7
C
45
8
A
47
9
B
55
10
B
55
11
C
42
12
A
50
Y..
Y2. Y..
Y21 Y2.
Y21 = + ( ─ ) + ( ─ )
45 = 46 + 5 + error
46
variety
yield
variety effect
Y21 = mean + var.effect + error
552
plot
mean yield per variety
error = ‐ 6
yield per
mean
variety
yield
var.effect
error
2
A
41
45
46
‐1
‐4
4
A
42
45
46
‐1
‐3
8
A
47
45
46
‐1
2
12
A
50
45
46
‐1
5
1
B
45
51
46
5
‐6
5
B
49
51
46
5
‐2
9
B
55
51
46
5
4
10
B
55
51
46
5
4
3
C
40
42
46
‐4
‐2
6
C
41
42
46
‐4
‐1
7
C
45
42
46
‐4
3
11
C
42
42
46
‐4
0
552
552
0
0
46
198
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
40
obs.
41
42
43
44
45
46
van Leeuwen
47
48
49
50
observation=
41
mean + 46
+
‐4
─
variety effect +
‐1
error
42
obs.
46
+
‐3
─
‐1
obs.
47
46
+
2
‐1
─
mean yield
yield
total variety effect
error
deviation SSvar.
SSerror SStotal
41
46
‐1
‐4
‐5
1
16
25
42
46
‐1
‐3
‐4
1
9
16
47
46
‐1
2
1
1
4
1
50
46
‐1
5
4
1
25
16
45
46
5
‐6
‐1
25
36
1
49
46
5
‐2
3
25
4
9
55
46
5
4
9
25
16
81
55
46
5
4
9
25
16
81
40
46
‐4
‐2
‐6
16
4
36
41
46
‐4
‐1
‐5
16
1
25
45
46
‐4
3
‐1
16
9
1
42
46
‐4
0
‐4
16
0
16
552
0
0
0
168
140
308
46
anova‐yield.xls
199
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
van Leeuwen
F in Table A.6. with df = 2,9
critical value = 4.26
F is significant
SS
df
MS
F
variety
168
2
84
error
total
140
308
9
11
15,56
P
5,4
0,029
how can you decrease the error variance even more?
(lower error variance more significance
by removing out of the error variance the variance which has a clear cause (e.g. variation in the field)
variation, which is inevitable in experimental fields
can be divided into components by using a block design in the experiment
variety i
Randomized complete block design
block j
200
block1
B
A
C
block2
A
B
C
block3
C
A
B
block4
B
C
A
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
van Leeuwen
variety test with onions, yield in kg/plot
Yij = Y.. + var.effi + block.effj + errorij
Yi.
Y..
variety effect = ─ block effect = Y. j ─ Y..
error = Yij ─ mean – var.effect – block effect
Y. j
Y..
Yi.
= Yij + ─ ─ plot
block
variety
yield
2
1
A
41
4
2
A
42
8
3
A
47
12
4
A
50
1
1
B
45
5
2
B
49
mean yield block
9
3
B
55
variety
10
4
B
55
A
41
42
47
50
45
3
1
C
40
B
45
49
55
55
51
6
2
C
41
C
40
41
45
42
42
7
3
C
45
mean
42
44
49
49
46
11
4
C
42
pivot table
1
552
46
201
2
3
4
mean
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
yield
yield
mean
block
variety
yield
plot
block
variety
yield
2
1
A
41
42
45
46
4
2
A
42
44
45
46
8
3
A
47
49
45
46
12
4
A
50
49
45
46
1
1
B
45
42
51
46
5
2
B
49
44
51
46
9
3
B
55
49
51
46
10
4
B
55
49
51
46
3
1
C
40
42
42
46
6
2
C
41
44
42
46
7
3
C
45
49
42
46
11
4
C
42
49
42
46
552
552
552
46
var‐eff
van Leeuwen
error
block‐
45‐46=‐1
0
42‐46=‐4
error = 41 + 46 – 42 – 45 = 0
yield
yield per block
yield per
mean
variety
yield
SSvar.
SSblock SSerror
block
variety
1
A
41
42
45
46
1
16
0
2
A
42
44
45
46
1
4
1
3
A
47
49
45
46
1
9
1
9
4
4
A
50
49
45
46
1
1
B
45
42
51
46
25
16
4
2
B
49
44
51
46
25
4
0
3
B
55
49
51
46
25
9
1
4
B
55
49
51
46
25
9
1
1
C
40
42
42
46
16
16
4
2
C
41
44
42
46
16
4
1
3
C
45
49
42
46
16
9
0
4
C
42
49
42
46
16
9
9
552
552
552
168
114
26
46
202
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
SSvar.
SSblock
168
van Leeuwen
SSerror SStotal
114
26
308
df
SS
df
MS
F
P
variety
block
error
total
variety effect: critical value (P=0.05) = variety‐effect is significant?
block effect: critical value (P=0.05) = block‐effect is significant?
Yij = mean + variety-effect + block-effect + error
yield
plot
per block variety block
yield
per variety
mean yield yield
variety block‐
‐effect effect
error
2
1
A
42
45
41
46
‐1
‐4
0
4
2
A
44
45
42
46
‐1
‐2
‐1
8
3
A
49
45
47
46
‐1
3
‐1
12
4
A
49
45
50
46
‐1
3
2
1
1
B
42
51
45
46
5
‐4
‐2
5
2
B
44
51
49
46
5
‐2
0
9
3
B
49
51
55
46
5
3
1
10
4
B
49
51
55
46
5
3
1
3
1
C
42
42
40
46
‐4
‐4
2
6
2
C
44
42
41
46
‐4
‐2
1
7
3
C
49
42
45
46
‐4
3
0
11
4
C
49
42
42
46
‐4
3
‐3
552
552
552
0
0
0
203
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
40
wrn
41
42
43
44
45
46
van Leeuwen
47
48
49
observation=
41
+
‐4
─
wrn
46
mean + ‐1
variety effect +
block effect +
error
42
46
+
‐1
─
‐2
‐1
wrn
47
+
46
3
‐1
─
mean yield yield
variety effect
block‐
effect
error
SSvariety SSblock
‐1
SSerror
41
46
‐1
‐4
0
1
16
0
42
46
‐1
‐2
‐1
1
4
1
47
46
‐1
3
‐1
1
9
1
50
46
‐1
3
2
1
9
4
45
46
5
‐4
‐2
25
16
4
49
46
5
‐2
0
25
4
0
55
46
5
3
1
25
9
1
55
46
5
3
1
25
9
1
40
46
‐4
‐4
2
16
16
4
41
46
‐4
‐2
1
16
4
1
45
46
‐4
3
0
16
9
0
42
46
‐4
3
‐3
16
9
9
0
0
0
168
114
26
552
46
204
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
van Leeuwen
Yij = Y.. + var.effi + block.effj + errorij
SS
df
MS
ras
blok
error
168
114
26
2
3
6
totaal
308
11
F
84
38
4,33
SS
19,38
8,77
df
P
0,002
0,013
MS
F
ras
168
2
84
error
140
9
15,56
totaal
308
11
P
5,4
0,029
variety effect is higher
significant with a block
design
SS
df
MS
F
P
ras
168
2
84
19,38
0,002
blok
114
3
38
8,77
0,013
error
26
6
4,33
308
11
totaal
205
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
yield
van Leeuwen
yield
block variety per block
per variety
yield
1
A
43
45
41
2
A
45
45
42
3
A
50
45
47
4
A
50
45
50
1
B
43
51
45
2
B
45
51
49
3
B
50
51
55
4
B
50
51
55
1
C
43
45
43
2
C
45
45
44
3
C
50
45
47
4
C
50
45
46
564
564
564
with other
observations
47,0
SS
ras
variety effect
not significant
with
randomized
design
SS
df
MS
F
96
2
48
error
136
9
15,11
totaal
232
11
P
3,18
0,090
variety-effect
significant with
randomized
block design
df
MS
F
P
ras
96
2
48
13,5
0,006
blok
115
3
38,22
10,75
0,0079
3,56
error
21
6
totaal
232
11
206
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
van Leeuwen
Anova in formulas
Source of
Sums of squares
variation
df
ras
t‐1
Definition
Computing formula
r * Yi. Y..
2
r‐1
t * Y. j Y..
(t‐1)(r‐1)
Total
t*r‐1=n‐1
Y
Y ─ t * r * Y
2
Y..
yield
mean
per var.
yield
2
2
ij
..
ij
t=3 varieties, r=4 blocks
SSvar.
Y = (45)2 = 2025
2
1.
1
A
41
42
45
46
2025
2
A
42
44
45
46
2025
3
A
47
49
45
46
2025
4
A
50
49
45
46
2025
1
B
45
42
51
46
2601
2
B
49
44
51
46
2601
3
B
55
49
51
46
2601
4
B
55
49
51
46
2601
1
C
40
42
42
46
1764
t * r * Y..
2
C
41
44
42
46
1764
(correction factor C)
3
C
45
49
42
46
1764
4
C
42
49
42
46
1764
552
552
552
46
2
j
ij
per bl. var. yield block
2
SSerror = SStotaal – SSras – SSblok
ij
yield
t * Y. j ─ t * r * Y..
2
2
i
j
Error
2
i
blok
r * Yi. ─ t * r * Y..
25560
25392
207
Y = (51)2 = 2601
2
2.
r * Yi.
2
i
= 4*(2025+2601+1764)=25560
2
=12*462=25392
SSvar. = SST =
25560‐25392=168
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
yield
per bl. var. yield block
yield
mean
per var.
yield
van Leeuwen
t=3 varieties, r=4 blocks
SSblock
Y = (42)2 = 1764
2
.1
1
A
41
42
45
46
1764
2
A
42
44
45
46
1936
3
A
47
49
45
46
2401
4
A
50
49
45
46
2401
1
B
45
42
51
46
1764
2
B
49
44
51
46
1936
3
B
55
49
51
46
2401
4
B
55
49
51
46
2401
1
C
40
42
42
46
1764
t * r * Y..
2
C
41
44
42
46
1936
(correction factor C)
3
C
45
49
42
46
2401
4
C
46
2401
42
49
42
552
552
552
25506
46
Y
2
.2
= (44)2 = 1936
t * Y. j
2
j
= 3*(1764+1936 +2401+2401) =25506
2
=12*462=25392
SSblock = 25506‐25392
=114
25392
yield
per bl. var. yield block
yield
mean
per var.
yield
t=3 varieties, r=4 blocks
SStotal
1
A
41
42
45
46
1681
2
A
42
44
45
46
1764
3
A
47
49
45
46
2209
4
A
50
49
45
46
2500
(Y11)2 = (41)2 = 1681
(Y12)2 = (42)2 = 1764
Y
2
ij
1
B
45
42
51
46
2025
2
B
49
44
51
46
2401
3
B
55
49
51
46
3025
4
B
55
49
51
46
3025
1
C
40
42
42
46
1600
t * r * Y..
2
C
41
44
42
46
1681
(correction factor C)
3
C
45
49
42
46
2025
4
C
42
49
42
46
1764
552
552
552
46
25700
25392
208
ij
= 1681+1764+….1764=25700
2
=12*462=25392
SStotal = 25700‐25392
=308
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
Source of
Sums of squares
variation
df
Uncorrected
Corrected
variety
3‐1=2
4‐1=3
25560
25506
25560 – 25392 = 168
25506 – 25392 = 114
block
Error
(3‐1)*
(4‐1)=6
3*4‐1 =11
Total
van Leeuwen
SS
SSerror = SStotal – SSvar.– SSblock
= 308 ‐ 168 ‐ 114 = 26
25700
df
25700 – 25392 = 308
MS
variety
168
2
block
114
error
26
total
308
11
F
P
84
19,38
0,002
3
38
8,77
0,013
6
4,33
Experiment to find out if there are differences between two or more treatments
Observations: variable Y
Starting point: variable is normally distributed
In a sample from this population (all possible values of Y),
95% of the observations is between µ ± 2σ
in small samples coincidence plays a role
209
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
If treatment is only applied to one experimental unit:
value of Y is determined by treatment and erroreffect
it is impossible to discriminate between the two effects
you need replicates
Experiment:
treatment = variety : 2 varieties, 4 replicates
We are interested in differences between varieties
Starting point: if the observations Y on a sample of a variety are normally distributed, the differences are also normally distributed
6
number
5
4
Reeks1
3
Reeks2
2
1
0
length cucumber
coincidence effect causes variance within samples
210
van Leeuwen
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
There are different methods to find out if differences between treatments are significant
significant means: differences are so big, taking into account the variances, that this difference is not caused by coincidence, but by the treatment effect
F‐test (ANOVA): are there any differences between t treatments?
t‐test: differences between two treatments
t‐test: comparing two independent samples, for example variety A and variety B
A
B
41
45
42
49
47
55
50
55
mean
45
51
13,5
18
s2
s=SD 3,67 4,24
d Y1 Y2
d
SED
AB
LSD = ?
211
df?
t=d/SED
t0,05;6;2tails
van Leeuwen
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
van Leeuwen
t‐test: comparing two independent samples, for example variety A and variety B
A
B
41
45
42
49
47
55
50
55
mean
45
51
2
13,5
18
s
s=SD 3,67 4,24
df?
d Y1 Y2
d
SED
t=d/SED
t0,05;6;2tails
AB
LSD = SED*t0,05;6;2tail
sd2=(sA2+sB2)/4=(13,5+18)/4 = 7,88
sd=√7,88 = 2,81 = SED
LSD = SED*t0,05;6;2tail= 2,81*2,14=6,82
If experiment with more than 2 treatments:
F‐test with ANOVA: is the treatment effect significant?
A
41
42
47
50
B
45
49
55
55
C
40
41
45
42
ANOVA
SS
variety 168
error 140
df
MS
2
84
9 15,56
F
P
5,4 0,029
After ANOVA and if treatmenteffect is significant: MSerror is sample variance and can be used to calculate SED, which can be used to calculate LSD for the total experiment.
Differences can be calculated pairwise and compared with this LSD
F‐test – protected LSD
AB
AC
BC
212
d
6
3
9
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
SS
df
MS
F
P
variety
168
2
84
19,38
0,002
block
114
3
38
8,77
0,013
4,33
error
26
6
total
308
11
van Leeuwen
If blocks are used MSerror is smaller
After the general question if the treatment effect (or blockeffect is significant, pairwise comparisons can be made
Standard Deviation from the Anova table is used:
Variance(error) = 4,33, SD = √4,33 = 2,08
difference between variety 1 and 2
t12
testing parameter:
d
SED
Y1 Y2
1 1
SD
n1 n2
critical value from t‐
table A.3:
t(0,05;df)
df = dferror
SS
df
MS
F
P
variety
168
2
84
19,38
0,002
block
114
3
38
8,77
0,013
4,33
error
26
6
total
308
11
LSD for variety differences?
Standard Error of the difference: SED
SED SD
n = 4 =replicates per variety
1
1
2
SD
n1 n2
n
df = df(error) = 6
SED = √4,33*√ (2/4) = 1,47
This method is more precise as you use the standard error of the total experiment
t(0,05;6) = 2,45
LSD = tvalue* SED = 2,45 * 1,47 = 3,6
213
Lecture 1.5.2
Analysis of Variance (CRD, RCB)
SS
df
MS
F
P
ras
168
2
84
19,38
0,002
blok
114
3
38
8,77
0,013
26
6
4,33
308
11
error
totaal
van Leeuwen
We can also use the standard deviation to calculate confidence intervals of the mean per variety or block
Standard Deviation from the Anova‐table: SD = √4,33 = 2,08
n = number of observations per variety or block
1
SE SD
n
t(0,05;df) from table
95% confidence interval =
X
df = dferror
variety A: 45 ± 2,45*1,04
± t(0,05;df) * SE
t(0,05;6) = 2,45
SE=2,08/√4=1,04
42,5 < < 47,5
XA
In a big experiment (for example 20 varieties) 190 pairwise comparisons are possible
190 t‐tests, each with a probability of 5% of type I error
(5% of tests are significant, while H0 is true)
5% x 190 = 9,5 9 or 10 false rejections of H0.
For that reason alternative t‐tests are used, which make a correction for these mistakes:
Bonferoni
Tukey
214
Table A-4.
Normal Distribution. This table contains the cumulative probabilities
exceeding the z-values under the right-hand side of a normal distribution
curve. The z-values are shown in the first column up to one digit after the
decimal point and, in the top row, for the second digit after the decimal
point.
X−μ
z=
σ
z
.0
.1
.2
.3
.4
.5
.6
.7
.8
.9
1.0
1.1
1.2
1.3
1.4
1.5
1.6
1.7
1.8
1.9
2.0
2.1
2.2
2.3
2.4
2.5
2.6
2.7
2.8
2.9
3.0
3.1
3.2
3.3
.00
.500
.460
.421
.382
.345
.309
.274
.242
.212
.184
.159
.136
.115
.097
.081
.067
.055
.045
.036
.029
.023
.018
.014
.011
.008
.006
.005
.003
.003
.002
.001
.001
.001
.000
.01
.496
.456
.417
.378
.341
.305
.271
.239
.209
.181
.156
.133
.113
.095
.079
.066
.054
.044
.035
.028
.022
.017
.014
.010
.008
.006
.005
.003
.002
.002
.001
.001
.001
.000
.02
.492
.452
.413
.374
.337
.302
.268
.236
.206
.179
.154
.131
.111
.093
.078
.064
.053
.043
.023
.027
.022
.017
.013
.010
.008
.006
.004
.003
.002
.002
.001
.001
.001
.000
.03
.488
.448
.409
.371
.334
.298
.264
.233
.203
.176
.152
.129
.109
.092
.076
.063
.052
.042
.034
.027
.021
.017
.013
.010
.008
.006
.004
.003
.002
.002
.001
.001
.001
.000
.04
.484
.444
.405
.367
.330
.295
.261
.230
.200
.174
.149
.127
.107
.090
.075
.062
.051
.041
.033
.026
.021
.016
.013
.010
.007
.006
.004
.003
.002
.002
.001
.001
.001
.000
215
.05
.480
.440
.401
.363
.326
.291
.258
.227
.198
.171
.147
.125
.106
.089
.074
.061
.049
.040
.032
.026
.020
.016
.012
.009
.007
.005
.004
.003
.002
.002
.001
.001
.001
.000
.06
.476
.436
.397
.359
.323
.288
.255
.224
.195
.169
.145
.123
.104
.087
.072
.059
.048
.039
.031
.025
.020
.015
.012
.009
.007
.005
.004
.003
.002
.002
.001
.001
.001
.000
.07
.472
.433
.394
.356
.319
.284
.251
.221
.192
.166
.142
.121
.102
.085
.071
.058
.047
.038
.031
.024
.019
.015
.012
.009
.007
.005
.004
.003
.002
.001
.001
.001
.001
.000
.08
.468
.429
.390
.352
.316
.281
.248
.218
.189
.164
.140
.119
.100
.084
.069
.057
.046
.038
.030
.024
.019
.015
.011
.009
.007
.005
.004
.003
.002
.001
.001
.001
.001
.000
.09
.464
.425
.386
.348
.312
.278
.245
.215
.187
.161
.138
.117
.099
.082
.068
.056
.046
.037
.029
.023
.018
.014
.011
.008
.006
.005
.004
.003
.002
.001
.001
.001
.001
.000
Table A-5.
df
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
Values of Chi-Square. This table is used to find values of chisquare corresponding to selected probabilities. Each number in
this table is a chi-square value for an indicated degree of freedom
(left column). Each number (i.e., .99, .95, ...) in the top row of the
table represents a probabiltiy of obtaining a value as large or larger
than the corresponding chi-square value in the table.
.99
0.0002
0.020
0.115
0.297
0.554
0.872
1.239
1.646
2.088
2.558
3.053
3.571
4.107
4.660
5.229
5.812
6.408
7.015
7.633
8.260
8.897
9.542
10.196
10.856
11.524
12.198
12.879
13.565
14.256
14.953
Probability of obtaining a value as large or larger
.95
.90
.50
.10
.05
.01
6.635
3.841
2.706
0.455
0.016
0.004
9.210
5.991
4.605
1.386
0.211
0.103
7.815 11.345
6.251
2.366
0.584
0.352
9.488 13.277
7.779
3.357
1.064
0.711
9.236 11.070 15.086
4.351
1.610
1.145
5.348 10.645 12.592 16.812
2.204
1.635
6.346 12.017 14.067 18.475
2.833
2.167
7.344 13.362 15.507 20.090
3.490
2.733
8.343 14.684 16.919 21.666
4.168
3.325
9.342 15.987 18.307 23.209
4.865
3.940
5.578 10.341 17.275 19.675 24.725
4.575
6.304 11.340 18.549 21.026 26.217
5.226
7.042 12.340 19.812 22.362 27.688
5.892
7.790 13.339 21.064 23.685 29.141
6.571
8.547 14.339 22.307 24.996 30.578
7.261
9.312 15.338 23.542 26.296 32.000
7.962
8.672 10.085 16.338 24.769 27.587 33.409
9.390 10.865 17.338 25.989 28.869 34.805
10.117 11.651 18.338 27.204 30.144 36.191
10.851 12.443 19.337 28.412 31.410 37.566
11.591 13.240 20.337 29.615 32.671 38.932
12.338 14.041 21.337 30.813 33.924 40.289
13.091 14.848 22.337 32.007 35.172 41.638
13.848 15.659 23.337 33.196 36.415 43.980
14.611 16.473 24.337 34.382 37.652 44.314
15.379 17.292 25.336 35.563 38.885 45.642
16.151 18.114 26.336 36.741 40.113 46.963
16.928 18.939 27.336 37.916 41.337 48.278
49.588
3.557
17.708 19.768 28.336 39.087
18.493 20.599 29.336 40.256 43.773 50.892
216
.001
10.828
13.816
16.266
18.467
20.515
22.458
24.322
26.124
27.877
29.588
31.264
32.909
34.528
36.123
37.697
39.252
40.790
42.312
43.820
45.315
46.797
48.268
49.728
51.179
52.620
54.052
55.476
56.892
58.301
59.703
Table A-6. Values of the student t-distribution. The values in this table are Student tvalues which correspond to the indicated cumulative probabilities
(on the right-hand side under a t-distribution curve) and to the
indicated degrees of freedom.
df
1
2
3
4
5
6
7
8
9
10
.25
1.000
0.816
0.765
0.741
0.727
0.718
0.711
0.706
0.703
0.700
.20
1.376
1.061
0.978
0.941
0.920
0.906
0.896
0.889
0.883
0.879
Probability of ofbtaining a larger value of t
.15
.10
.05
.025
.01
.005
1.963 3.078 6.314 12.706 31.821 63.657
4.303 6.965 9.925
1.386 1.886 2.920
3.182 4.451 5.841
1.250 1.638 2.353
2.776 3.747 4.604
1.190 1.533 2.132
2.571 3.365 4.032
1.156 1.476 2.015
2.447 3.143 3.707
1.134 1.440 1.943
2.365 2.998 3.499
1.119 1.415 1.895
2.306 2.896 3.355
1.108 1.397 1.860
2.262 2.821 3.250
1.100 1.383 1.833
2.228 2.764 3.169
1.093 1.372 1.812
.0005
636.619
31.599
12.924
8.610
6.869
5.959
5.408
5.041
4.781
4.587
11
12
13
14
15
16
17
18
19
20
0.697
0.695
0.694
0.692
0.691
0.690
0.689
0.688
0.688
0.687
0.876
0.873
0.870
0.868
0.866
0.865
0.863
0.862
0.861
0.860
1.088
1.083
1.079
1.076
1.074
1.071
1.069
1.067
1.066
1.064
1.363
1.356
1.350
1.345
1.341
1.337
1.333
1.330
1.328
1.325
1.796
1.782
1.771
1.761
1.753
1.746
1.740
1.734
1.729
1.725
2.201
2.179
2.160
2.145
2.131
2.120
2.110
2.101
2.093
2.086
2.718
2.681
2.650
2.624
2.602
2.583
2.567
2.552
2.539
2.528
3.106
3.055
3.012
2.977
2.947
2.921
2.898
2.878
2.861
2.845
4.437
4.318
4.221
4.140
4.073
4.015
3.965
9.922
3.883
3.850
21
22
23
24
25
26
27
28
29
30
0.686
0.686
0.685
0.685
0.684
0.684
0.684
0.683
0.683
0.683
0.859
0.858
0.858
0.857
0.856
0.856
0.855
0.855
0.854
0.854
1.063
1.061
1.060
1.059
1.058
1.058
1.057
1.056
1.055
1.055
1.323
1.321
1.319
1.318
1.316
1.315
1.314
1.313
1.311
1.310
1.721
1.717
1.714
1.711
1.708
1.706
1.703
1.701
1.699
1.697
2.080
2.074
2.069
2.064
2.060
2.056
2.052
2.048
2.045
2.042
2.518
2.508
2.500
2.492
2.485
2.479
2.473
2.467
2.462
2.457
2.831
2.819
2.807
2.797
2.787
2.779
2.771
2.763
2.756
2.750
3.819
3.792
3.768
3.745
3.725
3.707
3.690
3.674
3.659
3.646
40
∞
0.681
0.674
0.851
0.842
1.050
1.036
1.303
1.282
1.684
1.645
2.021
1.960
2.423
2.326
2.704
2.576
3.551
3.291
217
Table A-7. Values of the F-Distribution - 10%, 5% and 1% level. This table is used to find F-values at three probability
levels (.10, .05, .01) in the right tail of the F-distribution.
df for
denom
1
1
1
2
2
2
3
3
3
4
4
4
5
5
5
6
6
6
7
7
7
8
8
P
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
1
39.9
161
4052
8.52
18.51
98.50
5.54
10.13
34.12
4.54
7.71
21.20
4.06
6.61
16.26
3.78
5.99
13.75
3.59
5.59
12.25
3.46
5.32
2
49.5
199
5000
9.00
19.00
99.00
5.46
9.55
30.82
4.32
6.94
18.00
3.78
5.79
13.27
3.46
5.14
10.92
3.26
4.74
9.55
3.11
4.46
3
53.6
216
5403
9.16
19.2
99.2
5.39
9.28
29.5
4.19
6.59
16.7
3.62
5.41
12.1
3.29
4.76
9.78
3.07
4.35
8.45
2.92
4.07
4
55.8
225
5625
9.24
19.2
99.2
5.34
9.12
28.7
4.11
6.39
16.0
3.52
5.19
11.4
3.18
4.53
9.15
2.96
4.12
7.85
2.81
3.84
Degrees of Freedom for Numerator
5
6
7
8
9
10
60.2
59.9
59.4
58.9
58.2
57.2
242
241
239
237
234
230
5764 5859 5928 5981 6022 6056
9.39
9.38
9.37
9.35
9.33
9.29
19.4
19.4
19.4
19.4
19.3
19.3
99.4
99.4
99.4
99.4
99.3
99.3
5.23
5.24
5.25
5.27
5.28
5.31
8.79
8.81
8.85
8.89
8.94
9.01
27.2
27.3
27.5
27.9
27.9
28.2
3.92
3.94
3.95
3.98
4.01
4.05
5.96
6.00
6.04
6.09
6.16
6.26
14.5
14.7
14.8
15.0
15.2
15.5
3.30
3.32
3.34
3.37
3.40
3.45
4.74
4.77
4.82
4.88
4.95
5.05
10.1
10.2
10.3
10.5
10.7
11.0
2.94
2.96
2.98
3.01
3.05
3.11
4.06
4.10
4.15
4.21
4.28
4.39
7.87
7.98
8.10
8.26
8.47
8.75
2.70
2.72
2.75
2.78
2.83
2.88
3.64
3.68
3.73
3.79
3.87
3.97
6.62
6.72
6.84
6.99
7.19
7.46
2.73
2.67
2.62
2.59
2.56
2.54
3.69
3.58
3.50
3.44
3.39
3.35
218
12
60.7
244
6106
9.41
19.4
99.4
5.22
8.74
27.1
3.90
5.91
14.4
3.27
4.68
9.9
2.90
4.00
7.72
2.67
3.57
6.47
2.50
3.28
14
61.1
245
6143
9.42
19.4
99.4
5.20
8.71
26.9
3.88
5.87
14.2
3.25
4.64
9.8
2.88
3.96
7.60
2.64
3.53
6.36
2.48
3.24
16
61.3
246
6170
9.43
19.43
99.4
5.20
8.69
26.8
3.86
5.84
14.2
3.23
4.60
9.7
2.86
3.92
7.52
2.62
3.49
6.28
2.45
3.20
18
61.6
247
6192
9.44
19.4
99.4
5.19
8.67
26.8
3.85
5.82
14.1
3.22
4.58
9.6
2.85
3.90
7.45
2.61
3.47
6.21
2.44
3.17
df for
denom
8
9
9
9
10
10
10
11
11
11
12
12
12
13
13
13
14
14
14
15
15
15
16
16
16
17
17
17
P
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
1
11.26
3.36
5.12
10.56
3.29
4.96
10.04
3.23
4.84
9.65
3.18
4.75
9.33
3.14
4.67
9.07
3.10
4.60
8.86
3.07
4.54
8.68
3.05
4.49
8.53
3.03
4.45
8.40
2
8.65
3.01
4.26
8.02
2.92
4.10
7.56
2.86
3.98
7.21
2.81
3.89
6.93
2.76
3.81
6.70
2.73
3.74
6.51
2.70
3.68
6.36
2.67
3.63
6.23
2.64
3.59
6.11
3
7.59
2.81
3.86
6.99
2.73
3.71
6.55
2.66
3.59
6.22
2.61
3.49
5.95
2.56
3.41
5.74
2.52
3.34
5.56
2.49
3.29
5.42
2.46
3.24
5.29
2.44
3.20
5.18
4
7.01
2.69
3.63
6.42
2.61
3.48
5.99
2.54
3.36
5.67
2.48
3.26
5.41
2.43
3.18
5.21
2.39
3.11
5.04
2.36
3.06
4.89
2.33
3.01
4.77
2.31
2.96
4.67
5
6.63
2.61
3.48
6.06
2.52
3.33
5.64
2.45
3.20
5.32
2.39
3.11
5.06
2.35
3.03
4.86
2.31
2.96
4.69
2.27
2.90
4.56
2.24
2.85
4.44
2.22
2.81
4.34
Degrees of Freedom for Numerator
6
7
8
9
6.37
6.18
6.03
5.91
2.44
2.47
2.51
2.55
3.18
3.23
3.29
3.37
5.35
5.47
5.61
5.80
2.35
2.38
2.41
2.46
3.02
3.07
3.14
3.22
4.94
5.06
5.20
5.39
2.27
2.30
2.34
2.39
2.90
2.95
3.01
3.09
4.63
4.74
4.89
5.07
2.21
2.24
2.28
2.33
2.80
2.85
2.91
3.00
4.39
4.50
4.64
4.82
2.16
2.20
2.23
2.28
2.71
2.77
2.83
2.92
4.19
4.30
4.44
4.62
2.12
2.15
2.19
2.24
2.65
2.70
2.76
2.85
4.03
4.14
4.28
4.46
2.21
2.09
2.12
2.16
2.59
2.64
2.71
2.79
3.89
4.00
4.14
4.32
2.06
2.09
2.13
2.18
2.54
2.59
2.66
2.74
3.78
3.89
4.03
4.20
2.03
2.06
2.10
2.15
2.49
2.55
2.61
2.70
3.68
3.79
3.93
4.10
219
10
5.81
2.42
3.14
5.26
2.32
2.98
4.85
2.25
2.85
4.54
2.19
2.75
4.30
2.14
2.67
4.10
2.10
2.60
3.94
2.06
2.54
3.80
2.03
2.49
3.69
2.00
2.45
3.59
12
5.67
2.38
3.07
5.11
2.28
2.91
4.71
2.21
2.79
4.40
2.15
2.69
4.16
2.10
2.60
3.96
2.05
2.53
3.80
2.02
2.48
3.67
1.99
2.42
3.55
1.96
2.38
3.46
14
5.56
2.35
3.03
5.01
2.26
2.86
4.60
2.18
2.74
4.29
2.12
2.64
4.05
2.07
2.55
3.86
2.02
2.48
3.70
1.99
2.42
3.56
1.95
2.37
3.45
1.93
2.33
3.35
16
5.48
2.33
2.99
4.92
2.23
2.83
4.52
2.16
2.70
4.21
2.09
2.60
3.97
2.04
2.51
3.78
2.00
2.44
3.62
1.96
2.38
3.49
1.93
2.33
3.37
1.90
2.29
3.27
18
5.41
2.31
2.96
4.86
2.22
2.80
4.46
2.14
2.67
4.15
2.08
2.57
3.91
2.02
2.48
3.72
1.98
2.41
3.56
1.94
2.35
3.42
1.91
2.30
3.31
1.88
2.26
3.21
df for
denom
18
18
18
19
19
19
20
20
20
22
22
22
24
24
24
26
26
26
28
28
28
30
30
30
35
35
35
P
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
1
3.01
4.41
8.29
2.99
4.38
8.18
2.97
4.35
8.10
2.95
4.30
7.95
2.93
4.26
7.82
2.91
4.23
7.72
2.89
4.20
7.64
2.88
4.17
7.56
2.85
4.12
7.42
2
2.62
3.55
6.01
2.61
3.52
5.93
2.59
2.49
5.85
2.56
3.44
5.72
2.54
3.40
5.61
2.52
3.37
5.53
2.50
3.34
5.45
2.49
3.32
5.39
2.46
3.27
5.27
3
2.42
3.16
5.09
2.40
3.13
5.01
2.38
3.10
4.94
2.35
3.05
4.82
2.33
3.01
4.72
2.31
2.98
4.64
2.29
2.95
4.57
2.28
2.92
4.51
2.25
2.87
4.40
4
2.29
2.93
4.58
2.27
2.90
4.50
2.25
2.87
4.43
2.22
2.82
4.31
2.19
2.78
4.22
2.17
2.74
4.14
2.16
2.71
4.07
2.14
2.69
4.02
2.11
2.64
3.91
5
2.20
2.77
4.25
2.18
2.74
4.17
2.16
2.71
4.10
2.13
2.66
3.99
2.10
2.62
3.90
2.08
2.59
3.82
2.06
2.56
3.75
2.05
2.53
3.70
2.02
2.49
3.59
Degrees of Freedom for Numerator
6
7
8
9
2.00
2.04
2.08
2.13
2.46
2.51
2.58
2.66
3.60
3.71
3.84
4.01
1.98
2.02
2.06
2.11
2.42
2.48
2.54
2.63
3.52
3.63
3.77
3.94
1.96
2.00
2.04
2.09
2.39
2.45
2.51
2.60
3.46
3.56
3.70
3.87
1.93
1.97
2.01
2.06
2.34
2.40
2.46
2.55
3.35
3.45
3.59
3.76
1.91
1.94
1.98
2.04
2.30
2.36
2.42
2.51
3.26
3.36
3.50
3.67
1.96
2.01
1.88
1.92
2.39
2.47
2.27
2.32
3.42
3.59
3.18
3.29
1.87
2.00
1.90
1.94
2.24
2.29
2.36
2.45
3.12
3.23
3.36
3.53
1.98
1.93
1.88
1.85
2.42
2.33
2.27
2.21
3.47
3.30
3.17
3.07
1.82
1.85
1.90
1.95
2.16
2.22
2.29
2.37
2.96
3.07
3.20
3.37
220
10
1.98
2.41
3.51
1.96
2.38
3.43
1.94
2.35
3.37
1.90
2.30
3.26
1.88
2.25
3.17
1.86
2.22
3.09
1.84
2.19
3.03
1.82
2.16
2.98
1.79
2.11
2.88
12
1.93
2.34
3.37
1.91
2.31
3.30
1.89
2.28
3.23
1.86
2.23
3.12
1.83
2.18
3.03
1.81
2.15
2.96
1.79
2.12
2.90
1.77
2.98
2.84
1.74
2.04
2.74
14
1.90
2.29
3.27
1.88
2.26
3.19
1.86
2.22
3.13
1.83
2.17
3.02
1.80
2.13
2.93
1.77
2.09
2.86
1.75
2.06
2.79
1.74
2.04
2.74
1.70
1.99
2.64
16
1.87
2.25
3.19
1.85
2.21
3.12
1.83
2.18
3.05
1.80
2.13
2.94
1.77
2.09
2.85
1.75
2.05
2.78
1.73
2.02
2.72
1.71
1.99
2.66
1.67
1.94
2.56
18
1.85
2.22
3.13
1.83
2.18
3.05
1.81
2.15
2.99
1.78
2.10
2.88
1.75
2.05
2.79
1.72
2.02
2.72
1.70
1.99
2.65
1.69
1.96
2.60
1.65
1.91
2.50
df for
denom
40
40
40
45
45
45
50
50
50
60
60
60
∞
∞
∞
P
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
.10
.05
.01
1
2.84
4.08
7.31
2.82
4.06
7.23
2.81
4.03
7.17
2.79
4.00
7.08
2.71
3.84
6.63
2
2.44
3.23
5.18
2.42
3.20
5.11
2.41
3.18
5.06
2.39
3.15
4.98
2.30
3.00
4.61
3
2.23
2.84
4.31
2.21
2.81
4.25
2.20
2.79
4.20
2.18
2.76
4.13
2.06
2.60
3.78
4
2.09
2.61
3.83
2.07
2.58
3.77
2.06
2.56
3.72
2.04
2.53
3.65
1.94
2.37
3.32
5
2.00
2.45
3.51
1.98
2.42
3.45
1.97
2.40
3.41
1.95
2.37
3.34
1.85
2.21
3.02
Degrees of Freedom for Numerator
6
7
8
9
1.79
1.83
1.87
1.93
2.12
2.18
2.25
2.34
2.89
2.99
3.12
3.29
1.77
1.81
1.85
1.91
2.10
2.15
2.22
1.31
2.83
2.94
3.07
3.23
1.76
1.80
1.84
1.90
2.07
2.13
2.20
2.29
2.78
2.89
3.02
3.19
1.74
1.77
1.82
1.87
2.04
2.10
2.17
2.25
2.72
2.82
2.95
3.12
1.63
1.67
1.72
1.77
1.88
1.94
2.01
2.10
2.41
2.51
2.64
2.80
221
10
1.76
2.08
2.80
1.74
2.05
2.74
1.73
2.03
2.70
1.71
1.99
2.63
1.60
1.83
2.32
12
1.71
2.00
2.66
1.70
1.97
2.61
1.68
1.95
2.56
1.66
1.92
2.50
1.55
1.75
2.18
14
1.68
1.95
2.56
1.66
1.92
2.51
1.64
1.89
2.36
1.62
1.86
2.39
1.50
1.69
2.06
16
1.65
1.90
2.48
1.63
1.87
2.43
1.61
1.85
2.38
1.59
1.82
2.31
1.47
1.64
1.99
18
1.62
1.87
2.42
1.60
1.84
2.36
1.59
1.81
2.32
1.56
1.78
2.25
1.44
1.60
1.93