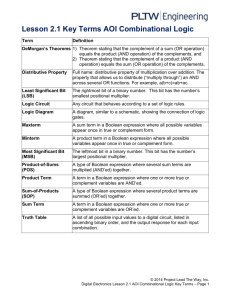
Optimal State Estimation for Boolean Dynamical Systems
advertisement

Optimal State Estimation for Boolean Dynamical
Systems using a Boolean Kalman Smoother
The 3rd IEEE Global Conference on Signal & Information Processing
Mahdi Imani and Ulisses Braga-Neto
Department of Electrical Engineering
Texas A&M University
College Station, Texas 77843
December 16, 2015
Introduction
Outline I
1
Introduction
2
Partially-Observable Boolean Dynamical Systems
3
Boolean Kalman Filter
4
Boolean Kalman Smoother
5
Numerical Experiments with RNA-Seq Count Data
6
Conclusions and Future Work
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
2 / 24
Introduction
Introduction
Boolean networks have emerged as an effective model of the
dynamical behavior of regulatory circuits consisting of bi-stable
components.
In the Boolean network model, the transcriptional state of each gene
is represented by 0 (OFF) or 1 (ON), and the relationship among
genes is described by logical gates updated and observed at discrete
time intervals.
This model has been successful in accurately modeling the dynamics
of the cell cycle in the Drosophila fruit fly, in the Saccharomyces
cerevisiae yeast, as well as the mammalian cell cycle.
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
3 / 24
Partially-Observable Boolean Dynamical Systems
Outline I
1
Introduction
2
Partially-Observable Boolean Dynamical Systems
3
Boolean Kalman Filter
4
Boolean Kalman Smoother
5
Numerical Experiments with RNA-Seq Count Data
6
Conclusions and Future Work
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
4 / 24
Partially-Observable Boolean Dynamical Systems
Partially-Observable Boolean Dynamical Systems
Boolean State Transition Model: there is uncertainty in state
transition. The sequence of state vectors {Xk ; k = 0, 1, . . .} is a
Markov stochastic process, called the state process, specified by
Xk = f (Xk−1 , uk−1 ) ⊕ nk ,
(1)
uk−1 and f are the input and network function, respectively, whereas
{nk ; k = 1, 2, . . .} is a white noise process.
Observation Model: In most real-world applications, the system state
is only partially observable, and distortion is introduced in the
observations by environmental or sensor noise:
Yk = h (Xk , vk ) ,
(2)
where vk is the observation noise.
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
5 / 24
Boolean Kalman Filter
Outline I
1
Introduction
2
Partially-Observable Boolean Dynamical Systems
3
Boolean Kalman Filter
4
Boolean Kalman Smoother
5
Numerical Experiments with RNA-Seq Count Data
6
Conclusions and Future Work
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
6 / 24
Boolean Kalman Filter
Boolean Kalman Filter
The Boolean Kalman Filter (BKF) is the recursive minimum mean-square
error (MMSE) state estimator X̂k = h(Y1 , . . . , Yk ) of the state Xk ,
according to the (conditional) mean-square error (MSE):
h
i
MSE(Y1 , . . . , Yk ) = E ||X̂k − Xk ||2 | Yk , . . . , Y1
(3)
Theorem
(Boolean Kalman Filter.) The optimal minimum MSE estimator X̂k of
the state Xk given the observations Y1 , . . . , Yk up to time k, is given by
X̂k = E [Xk | Yk , . . . , Y1 ] ,
(4)
where v(i) = Iv(i)>1/2 for i = 1, . . . , d.
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
7 / 24
Boolean Kalman Filter
Define the following distribution vectors of length 2d :
Πk|k (i) = P Xk = xi | Yk , . . . , Y1 ,
Πk|k−1 (i) = P Xk = xi | Yk−1 , . . . , Y1 ,
∆k|k (i) = P Yk+1 , . . . , YT | Xk = xi ,
∆k|k−1 (i) = P Yk , . . . , YT | Xk = xi ,
(5)
Prediction Matrix:
(Mk )ij = P(Xk = xi | Xk−1 = xj )
(6)
= P nk = xi ⊕ f(xj , uk−1 ) ,
Update Matrix:
(Tk )jj = P Yk | Xk = xj ,
(7)
d
Boolean States: A = [x1 ...x2 ]
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
8 / 24
Boolean Kalman Filter
Boolean Kalman Filter
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
9 / 24
Boolean Kalman Smoother
Outline I
1
Introduction
2
Partially-Observable Boolean Dynamical Systems
3
Boolean Kalman Filter
4
Boolean Kalman Smoother
5
Numerical Experiments with RNA-Seq Count Data
6
Conclusions and Future Work
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
10 / 24
Boolean Kalman Smoother
Boolean Kalman Smoother
The Boolean Kalman Smoother (BKS) is the minimum mean-square error
(MMSE) state estimator X̂S = g (Y1 , . . . , YT ) of the state XS , where
1 < S < T , according to the (conditional) mean-square error (MSE):
h
i
MSE(Y1 , . . . , YT ) = E ||X̂S − XS ||2 | YT , . . . , Y1
(8)
Theorem
(Boolean Kalman Smoother.) The optimal minimum MSE estimator
X̂S of the state XS given the observations Y1 , . . . , YT , where 1 < S < T ,
is given by
X̂S = E [XS | YT , . . . , Y1 ] ,
(9)
where v(i) = Iv(i)>1/2 for i = 1, . . . , d.
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
11 / 24
Boolean Kalman Smoother
Boolean Kalman Smoother
Boolean Kalman Smoother
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
12 / 24
Numerical Experiments with RNA-Seq Count Data
Outline I
1
Introduction
2
Partially-Observable Boolean Dynamical Systems
3
Boolean Kalman Filter
4
Boolean Kalman Smoother
5
Numerical Experiments with RNA-Seq Count Data
6
Conclusions and Future Work
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
13 / 24
Numerical Experiments with RNA-Seq Count Data
Parametric State Transition Model:
(
P
1,
aij (Xk−1 )j + bi + (uk−1 )i > 0
fi (Xk−1 , uk−1 ) =
Pj
0,
j aij (Xk−1 )j + bi + (uk−1 )i < 0
(10)
aij = +1 (Positive Regulation), aij = −1 (Negative Regulation), aij = 0 (No Regulation)
bi = +1/2 (Positively Biased), bi = −1/2 (Negatively Biased)
f
f
f
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
14 / 24
Numerical Experiments with RNA-Seq Count Data
RNA-Seq Observation Model:
1
RNA-seq Data
In this study, we choose to use a Poisson model for the number of
reads for each transcript:
P(Yki = m | λki ) = e −λki
λm
ki
,
m!
m = 0, 1, . . .
(11)
λki is the mean read count of transcript i at time k
log(λki ) = log(s) + µb ,
if Xki = 0 ,
log(λki ) = log(s) + µb + δi ,
if Xki = 1 .
(12)
s: sequencing depth,
µb > 0: Baseline expression,
δi > 0: Differential expression .
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
15 / 24
Numerical Experiments with RNA-Seq Count Data
Case Study: p53-MDM2 Negative Feedback Loop Pathway
1101
dna_dsb=0
(DNA damage)
1010
dna_dsb=1
(DNA damage)
1101
1100
dna dsb
0100
0110
1111
ATM
1011
p53
1011
1000
0101
0111
1110
0100
1010
0011
0101
0000
1001
1000
0001
Mdm2
Wip1
0010
0010
0010
1100
0001
1001
0000
0110
1110
0011
0111
(a)
Mahdi Imani, Ulisses Braga-Neto
(b)
GlobalSIP 2015
1111
(c)
December 16, 2015
16 / 24
Numerical Experiments with RNA-Seq Count Data
Average Performance of BKF and BKS.
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
17 / 24
Numerical Experiments with RNA-Seq Count Data
P=0.01
Average MSE
BKF
BKS
P=0.1
Average MSE
BKF
BKS
Time
Average MSE of BKF and BKS over 1000 runs.
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
18 / 24
Numerical Experiments with RNA-Seq Count Data
Case Study: Cell Cycle Regulatory Network
Rb
CycD
E2F
CycE
p27
CycB
Cdh1
CycA
Cdc20
Mahdi Imani, Ulisses Braga-Neto
UbcH10
GlobalSIP 2015
December 16, 2015
19 / 24
Numerical Experiments with RNA-Seq Count Data
State Transition
Original Trajectory
P=0.01
Estimated Trajectory (BKF)
Estimated Trajectory (BKS)
P=0.1
Time
Time
Time
Time
Estimated Trajectories by BKF and BKS.
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
20 / 24
Conclusions and Future Work
Outline I
1
Introduction
2
Partially-Observable Boolean Dynamical Systems
3
Boolean Kalman Filter
4
Boolean Kalman Smoother
5
Numerical Experiments with RNA-Seq Count Data
6
Conclusions and Future Work
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
21 / 24
Conclusions and Future Work
Conclusions and Future Work
We proposed a method for state estimation for Boolean dynamical
system observed though a single time series of noisy measurements
given the entire history of observations.
Future work includes:
Dealing with the network inference problem in the presence of batch
data.
Developing methods for discrete, continuous and mixed parameter
estimation.
Deriving efficient methods for large networks exploring sparsity, for
both state and parameter estimation.
Mahdi Imani, Ulisses Braga-Neto
GlobalSIP 2015
December 16, 2015
22 / 24
Thank you
Q&A