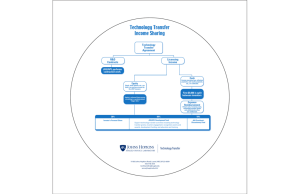
Project Summary - Johns Hopkins University
advertisement

Project Summary MRI: The Development of Data-Scope – A Multi-Petabyte Generic Data Analysis Environment for Science PI: Alexander Szalay, Co-Is: Kenneth Church, Charles Meneveau, Andreas Terzis, Scott Zeger The Data-Scope is a new scientific instrument, capable of ‘observing’ immense volumes of data from various scientific domains such as astronomy, fluid mechanics, and bioinformatics. Intellectual Merit: The nature of science is changing – new discoveries will emerge from the analysis of large amounts of complex data generated by our high-throughput instruments: this is Jim Gray’s “Fourth Paradigm” of scientific discovery. Virtual instruments (i.e., computers) generate equally large volumes of data – the sizes of the largest numerical simulations of nature today are on par with the experimental data sets. This data deluge is not simply a computational problem, but rather requires a new and holistic approach. We need to combine scalable algorithms and statistical methods with novel hardware and software mechanisms, such as deep integration of GPU computing with database indexing and fast spatial search capabilities. Today, scientists can easily tackle data-intensive problems at the 5-10TB scales: one can perform these analyses at a typical departmental computing facility. However, 50-100 TB are considerably more difficult to deal with and perhaps only ten universities in the world can analyze such data sets. Moving to the petabyte scale, there are less than a handful of places anywhere in the world that can address the challenge. At the same time there are many projects which are crossing over the 100TB boundary today. Astrophysics, High Energy Physics, Environmental Science, Computational Fluid Dynamics, Genomics, and Bioinformatics are all encountering data challenges in the several hundred TB range and beyond – even within the Johns Hopkins University. The large data sets are here, but we lack an integrated software and hardware infrastructure to analyze them! We propose to develop the Data-Scope, an instrument specifically designed to enable data analysis tasks that are simply not possible today. The instrument’s unprecedented capabilities combine approximately five Petabytes of storage with a sequential IO bandwidth close to 500GBytes/sec, and 600 Teraflops of GPU computing. The need to keep acquisition costs and power consumption low, while maintaining high performance and storage capacity introduces difficult tradeoffs. The Data-Scope will provide extreme data analysis performance over PB-scale datasets at the expense of generic features such as fault tolerance and ease of management. This is however acceptable since the Data-Scope is a research instrument rather than a traditional computational facility. Over the last decade we have demonstrated the ability to develop and operate data-intensive systems of increasing scale. Broader Impact: The data-intensive nature of science is becoming increasingly important. We face today a similar vacuum in our abilities to handle large data sets as the one from which the concept of the Beowulf cluster emerged in the 90s that eventually democratized high-performance computing. Many universities and scientific disciplines are now looking for a new template that will enable them to address PB-scale data analysis problems. In developing the Data-Scope, we can substantially strengthen the Nation’s expertise in data-intensive science. In order to accelerate the acceptance of the proposed approach we will collaborate with researchers across multiple disciplines and institutions nationwide (Los Alamos, Oak Ridge, UCSC, UW, STScI and UIC). The proposed instrument will also host public services on some of the largest data sets in astronomy and fluid mechanics, both observational and simulated. Students and postdoctoral fellows who will be involved in data-intensive research using the Data-Scope will acquire skills that will serve them well in their career as 21st century scientists. Partnerships: We have a strong industrial involvement and interest. We have been working with Microsoft Research and the SQL Server team for over a decade exploring ways to bring data-intensive computations as close to the data as possible. Microsoft has provided substantial funding to build the GrayWulf facility, the forerunner to the Data-Scope. We will continue to collaborate with Microsoft to advance innovations in data-intensive computing. NVIDIA is extremely interested in using GPUs in data intensive computations and in building data-balanced architectures and has recently awarded JHU a CUDA Research Center status. JHU is an active partner in the Open Cloud Consortium (OCC). The DataScope will use the 10Gbps OCC connectivity to move external large data sets into the instrument and will be linked to the rest of the OCC infrastructure. Project Description (a) Instrument Location The Data-Scope will be located in Room 448 at the Bloomberg Center for Physics and Astronomy. This room has adequate electricity and cooling to support 180kVA. The equipment currently residing there will be moved to a new location during the summer of 2010. The room is also instrumented with environmental sensors monitoring the temperature of each rack and the electricity of each circuit through a wireless sensor network connected to a real-time monitoring system. (b) Research Activities to be enabled Astrophysics, Physical Sciences Alex Szalay, Rosemary Wyse, Mark Robbins, Morris Swartz, Tamas Budavari, Ani Thakar, Brice Menard, Mark Neyrinck (JHU Physics and Astronomy), Robert Hanisch (Space Telescope Science Institute), Piero Madau, Joel Primack (UC Santa Cruz), Andrew Connolly (Univ. Washington), Salman Habib (Los Alamos), Robert Grossman (UIC) From 2010 JHU became the long term host of the Sloan Digital Sky Survey (SDSS) Archive [1], (Szalay, Thakar). This archive is one of astronomy’s most used facilities today. On the Data-Scope we can lay out the SDSS data multiple times and achieve orders of magnitude better performance for the whole astronomy community. The long term curation of the SDSS archive is one of the main thrusts for the NSFfunded Data Conservancy (Choudhury). The Data-Scope will enable new functionalities over the SDSS data that would perform complex computations over arbitrary subsets in a matter of seconds (Menard). The Virtual Astronomical Observatory [2] (VAO, directed by R. Hanisch provides the framework for discovering and accessing astronomical data from observatories and data centers. Perhaps the single most important VAO tool in the research community is its cross-matching service, OpenSkyQuery [3], built at JHU. Survey catalogs today contain 108 or more objects, while the next-generation telescopes, like LSST [4], Pan-STARRS [5] will soon produce orders of magnitude larger time-domain catalogs. Data sets this large can only be explored with a parallel computational instrument optimized for both I/O and CPU. The OpenSkyQuery, running on the Data-Scope (Budavari, Wilton) will improve performance for astronomical cross-matching by a factor of 10-100, helping the whole community. Access to large simulations has always been limited and awkward. The Data-Scope will host several of the world’s largest cosmological simulations [6,7,8] (Primack, Habib) and make them publicly accessible through interactive web services (like in turbulence). In particular, we will also combine observational data [9] and simulations of unprecedented size (300TB+) about the Milky Way (Wyse, Madau, Szalay). Our postdocs (Neyrinck) and graduate students will generate a suite of 500 realizations of a 1Gpc cosmological simulation, for multiple analyses of large scale structure. We will explore introducing arrays as native data types into databases to accelerate the analysis of structured data, in collaboration with Jose Blakeley (Microsoft), one of the Chief Architects of the SQL Server database. The total volume of the astrophysical simulation effort can easily exceed 500TB. JHU has been a member of the Open Cloud Consortium [10], led by R. Grossman (UIC). The OCC has a unique infrastructure, using its own high-speed networking protocols to transfer large data sets (20 min for a TB from Chicago to JHU). This framework enables us to import these large simulations directly from the National Supercomputer Centers (ORNL, NERSC), and also transfer the results to other institutions and NSF facilities (UCSD, UIUC, TeraGrid). The Data-Scope will also be available through the OCC. Detailed image simulations for LSST have been carried out [12] on the Google/IBM MR cluster (Connolly) but that system is reaching its resource limits, more storage and processing power is needed than what is available. The Data-Scope system can offer both, and the simulations can be expanded to 100TB. State of the art simulations of molecular dynamics on the largest HPC facilities use billion particle systems, representing about 1000 particles in each spatial direction, or about 300nm for atomic systems. The NSF MRI funded Graphics Processor Laboratory (NSF MRI-0923018, Acquisition of 100TF Graphics Processor Laboratory for Multiscale/Multiphysics Modeling, PI: Robbins, CoIs: Burns, Graham-Brady, Szalay, Darlymple) is enabling us to port the relevant codes to the GPU architecture. Integrating this system with the Data-Scope and using it to analyze thousands of configurations for billion particle simulations would enable dramatically better understanding of the mechanics of disordered systems and scaling behavior of avalanches as materials fail (Falk and Robbins). Saving the positions, velocities and accelerations of two such simulations requires 144TB, which is consistent with the size of the planned instrument, and would result in a dramatic breakthrough in the state of the art in this field. Data is already flowing in from the Large Hadron Collider (LHC). JHU has a 20TB local facility to analyze the data and produce simulated data (Swartz), which has already been outgrown. Having access to a 200TB revolving storage and fast network transfers from FNAL would dramatically impact the quality of the JHU contribution to the LHC effort. Turbulence, computational fluid mechanics Charles Meneveau (ME), Shiyi Chen (ME), Greg Eyink (AMS), Omar Knio (ME), Rajat Mittal (ME), Randal Burns (CS), Tony Darlymple (CE) The proposed Data-Scope will enable path-breaking research in fluid mechanics and turbulence. Constructing, handling, and utilizing large datasets in fluid dynamics has been identified at two NSF workshops [12,13] as a major pacing item for continued progress in the field. Scientific computing of multi-scale physical phenomena is covering increasingly wider ranges of spatial and temporal scales. Discretization and integration forward in time of the underlying partial differential equations constitute massive simulations that describe the evolution of physical variables (velocity, pressure, concentration fields) as function of time and location in the entire domain of interest. The prevailing approach has been that individual researchers perform large simulations that are analyzed during the computation, and only a small subset of time-steps are stored for subsequent, and by necessity more “static”, analysis. The majority of the time evolution is discarded. As a result, much of the computational effort is not utilized as well as it should. In fact, often the same simulations must be repeated after new questions arise that were not initially obvious. But many (or even most) breakthrough concepts cannot be anticipated in advance, as they will be motivated in part by output data and must then be tested against it. Thus a new paradigm is emerging that aims to create large and easily accessible databases that contain the full space-time history of simulated flows. The Data-Scope will provide the required storage and access for the analysis. The proposed effort in fluid mechanics and turbulence builds upon the accomplishments of two prior related projects: ITR-AST-0428325: Exploring the Lagrangian Structure of Complex Flows with 100 Terabyte Datasets (09/04 to 08/10; PIs: A. Szalay, E. Vishniac, R. Burns, S. Chen & G. Eyink), which used an MRI-funded cluster (funded through MRI-320907 DLMS: Acquisition of Instrumentation for a Digital Laboratory for Multi-Scale Science, PIs: Chen et al.). The cluster consists of a compute layer and a database layer, the JHU public turbulence database. It houses a 27 TB database that contains the entire time history of a 10243 mesh point pseudo-spectral DNS of forced isotropic turbulence [14,15]. 1024 time-steps have been stored, covering a full “large-eddy” turnover time. A database web service fulfills user requests for velocities, pressure, various space-derivatives of velocity and pressure, and interpolation functions (see http://turbulence.pha.jhu.edu). The 10244 isotropic turbulence database has been in operation continuously since it went online in 2008 and has been used extensively. Usage monitoring has shown that it has been accessed by over 160 separate IP’s and to date the total number of points queried exceeds 3.6 x 1010. Our prior research results using, or motivated by, the database are described in various publications [16-34]. For sample results from non-JHU users see e.g. papers [35,36]. The Data-Scope will enable us to build, analyze and provide publicly accessible datasets for other types of flows besides isotropic turbulence. We anticipate that over the next 2-4 years there will be about 5-7 new datasets created from very large CFD and turbulence simulations. The scale of the overall data will be in the 350-500 TB range. Anticipated topics include magneto-hydrodynamic turbulence, channel flow, atmospheric turbulence, combustion, compressible turbulence[37], cardiovascular and animal motion flows[38], free-surface flows, and propagation and quantification of uncertainty in model data[39,40]. Once these DBs are made accessible online, based on our experience with the isotropic turbulence DB, we anticipate that about 10 non-JHU research groups will regularly access each of the datasets. Hence, we expect about 50 external research groups (US and international) to profit from the instrument. Name and affiliation Science area A. Szalay, A. Thakar, S. Choudhury B.Menard +postdocs R Hanisch (STScI), T. Budavari, A. Thakar, Astronomy, data mgmt P. Madau (UCSC), R. Wyse, A. Szalay, 2 postdoc, 2 PhD st S. Habib (Los Alamos), J. Primack (UCSC) Astrophysics A. Connolly (UW)+postdocs M. Neyrinck, A. Szalay, 2 PhD students M. Robbins, M. Falk M. Swartz G. Eyink, E. Vishniac , R. Burns, 1 postdoc, 1 PhD st C. Meneveau, S. Chen, R. Burns, 1 PhD student O. Knio, C. Meneveau, R. Burns, 3 SANDIA, 2 PhD st C. Meneveau + 1 PhD st Astrophysics Astrophysics Astrophysics Virtual Astronomical Observatory Related project/agency TB SDSS archive services (NSF, Sloan Foundation) SDSS data analysis OpenSkyQuery, VO Spectrum, VO Footprint (NSF+NASA) 70 30 40 Via Lactea/The Milky Way Laboratory, (DOE, NSF) Public cosmology simulations (DOE,NASA) 300+ 100 50 Physics, Mat sci High Energy Phys. Turbulence, CFD, Astrophysics Turbulence, Mech. Eng. LSST imaging simulations Public access for 500 Gadget2 cosmological simulations Multiscale sims of avalanche Analysis of LHC data MHD database (NSF CDI) 144 250 100 Channel flow DB (NSF CDI) 80 Combustion, Energy, Mech Eng Env. Eng., atmospheric flow, wind energy. Chemically reacting flows with complex chemistry (DOE BES) Daily cycle of atmospheric boundary layer (NSF CBET) S. Chen, C. Meneveau + 1 PhD st Turbulence, Aerospace, propulsion Compressible turbulence (AFOSR, in planning) R. Mittal D. Valle + faculty + postdocs Genomics/Genotyping Cardiovascular & animal motion flows Epigenetic biomarkers, pathway Anal. (NCI,NIDCR,NLM) Improving communication in the ICU Multiple genome/whole exome 10-20 M. Ochs, Postdocs: E. Fertig and A. Favorov H. Lehmann Biomedical and biological fluid mech Systems biology, networks Biomedical informatics S. Yegnasubramanian, A. DeMarzo, W. Nelson, 4 Postdocs G. Steven Bova High throughput sequencing Ultra-high dim genomics correlations of very large datasets; (DoD, NCI, MD StemCell Fund) Distinctive genomic profiles of aggressive prostate cancer (DoD, pending) Correlation of multidimensional biological data (NSF+pending) Petascale Ocean Expt (NSF) 10-50 Astrophysics S. Wheelan, Postdoc: L. Mularoni T. Haine Correlation across highdimensional genomics datasets High throughput sequencing Env. Sci/Oceanography D. Waugh Env. Sci./Atmosphere B. Zaitchik Env. Sci./Hydrology Hugh Ellis Env. Sci./Air Quality 120 60-100 100 50 0.5/yr 9/yr 20-50 Chemistry-Climate Coupling (NSF,NASA) Regional climate analysis (NASA) Air Quality and Public Health (EPA, NSF) Table 1. Tabular representation of the major users of the Data-Scope Instrument and their data sizes 120 1050/yr 500 100 30 30 Bioinformatics Michael Ochs (Oncology, Biostatistics), Harold Lehmann (Public Health), Srinivasan Yegnasubramanian, Angelo DeMarzo, William Nelson (Oncology), G. Steven Bova (Pathology, Urology), Sarah Wheelan (Oncology, Biostatistics) Research activities that require or will be significantly accelerated by the Data-Scope are diverse and include high-dimensional biology, high-throughput sequencing, and algorithmic development to improve inference of biological function from multidimensional data. Harold Lehmann and colleagues have examined ICU activities and are striving for better integration of the massive amount of information produced daily in the unit. In general, they have been hampered by the inability to perform statistical analyses that use machine learning to find patterns, gaps, and warnings in patient courses and in team behavior. The volume of digital signals available from the many monitors applied to patients amounts to approximately 9TB per year in a modest-sized ICU. The proposed infrastructure would enable in-depth analysis to tackle quality and safety issues in healthcare–prediction of patient courses with and without interventions, and to include team activity in the mix. The researchers would include ICU and patient-safety researchers, computer scientists, statisticians, and their students. Srinivasan Yegnasubramanian, Angelo DeMarzo, and William Nelson jointly direct work on large, dynamic and high-dimensional datasets. In particular, they are working to characterize the genetics and epigenetics of normal hematopoietic and chronic myeloid leukemia stem cells as well as simultaneous genome-wide characterization of somatic alterations in imprinting, DNA methylation, and genomic copy number in prostate cancer. Dr. Yegnasubramanian, along with co-director Sarah J. Wheelan, M.D., Ph.D., also directs the Next Generation Sequencing Center, which is a shared resource for the entire genomics research community at the Sidney Kimmel Comprehensive Cancer Center as well as the JHU Schools of Medicine and Public Health at large. The center is expected to generate on the order of 100TB to 1PB of data per year using 4 ultra-high throughput Next Generation Sequencing Instruments. The capability to analyze these data in the Data-Scope, allowing integration and cross-cutting analyses of independently generated datasets will be of central importance. Michael Ochs directs several projects whose goal is to identify markers of cancer and cancer progression. The Data-Scope will allow Dr. Ochs to implement sophisticated probabilistic models, using known biology, data from many different measurement platforms, and ongoing experiments that will enable better prediction and management of disease. G. Steven Bova directs a large-scale effort in prostate cancer; many of the algorithms needed already exist but cannot operate as needed on his extremely large and diverse dataset, without an instrument such as the Data-Scope. Sarah Wheelan is working to develop methods for creating biologically relevant hypotheses from high throughput sequencing datasets. Such datasets typically bring several hundreds of millions of observations, and new algorithms are required to help biological investigators create the relevant and sophisticated queries that are possible with these data. By cross linking many different types of experiments across many different systems and using straightforward statistical techniques to survey correlations, new relationships can be uncovered. Climate and Environmental Science Hugh Ellis (DOGEE), Darryn Waugh, Tom Haine, Ben Zaitchik, Katalin Szlavecz (EPS) Several JHU faculty are performing high-resolution modeling and data assimilation of Earth's atmosphere, oceans, and climate system. The presence of the Data-Scope will allow multiple JHU users to access climate reanalysis data, satellite-derived datasets, and stored model output, and significantly accelerate the scientific analysis by permitting data exploration and visualization that would otherwise not be possible. The Data-Scope will also facilitate storage and analysis of large ensemble integrations, contributing to our ability to characterize uncertainty of the simulations and predictions. One example of the relevant research is the Petascale Arctic-Atlantic- Antarctic Experiment (PAVE) project (Haine). Under this project kilometer-resolution, planetary-scale, ocean and sea-ice simulations are being developed that will contain 20 billion grid cells, will require 10-40TB of memory, and will exploit petascale resources with between 100,000 and 1,000,000 processor cores. Local access to PAVE solutions, stored on the Data-Scope, will significantly accelerate the scientific analysis. Other examples include projects examining regional climate modeling with data assimilation (Zaitchik); the impact of air quality on public health in the US (Ellis); coupling between stratospheric chemistry and climate (Waugh); downscaled meteorological simulations of climate change-related health effects (Ellis). The simulations in these projects again produce large volumes of output, not only because of the required number of simulations and the need for fairly high spatial and temporal resolution, but also the large number of variables (e.g., large numbers of chemical species in air quality modeling together with numerous physical properties in meteorological simulation). The projects involving data assimilation also require run-time ingestion of large gridded datasets. In addition to these computer simulations the JHU wireless sensor networks (Szlavecz, Terzis) are providing in-situ monitoring of the soil’s contribution to the carbon cycle, and generate smaller (100 million records collected thus far) but quite complex data sets which need to be correlated and integrated with the large scale climate models and biological survey data. (c) Description of the Research Instrumentation and Needs Rationale for the Data-Scope The availability of large experimental datasets coupled with the potential to analyze them computationally is changing the way we do science [41,42]. In many cases however, our ability to acquire experimental data outpaces our ability to process them leading to the so-called data deluge [43]. This data deluge is the outcome of three converging trends: the recent availability of high throughput instruments (e.g., telescopes, high-energy particle accelerators, gene sequencing machines), increasingly larger disks to store the measurements, and ever faster CPUs to process them. Not only experimental data are growing at a rapid pace; the volume of data produced by computer simulations, used in virtually all scientific disciplines today, is increasing at an even faster rate. The reason is that intermediate simulation steps must also be preserved for future reuse as they represent substantial computational investments. The sheer volume of these datasets is only one of the challenges that scientists must confront. Data analyses in other disciplines (e.g., environmental sciences) must span thousands distinct datasets with incompatible formats and inconsistent metadata. Overall, dataset sizes follow a power law distribution and challenges abound at both extremes of this distribution. While improvements in computer hardware have enabled this data explosion, the performance of different architecture components increases at different rates. CPU performance has been doubling every 18 months, following Moore’s Law [44]. The capacity of disk drives is doubling at a similar rate, somewhat slower that the original Kryder’s Law prediction [45], driven by higher density platters. On the other hand, the disks’ rotational speed has changed little over the last ten years. The result of this divergence is that while sequential IO speeds increase with density, random IO speeds have changed only moderately. Due to the increasing difference between the sequential and random IO speeds of our disks, only sequential disk access is possible – if a 100TB computational problem requires mostly random access patterns, it cannot be done. Finally, network speeds, even in the data center, are unable to keep up with the doubling of the data sizes [46]. Said differently, with petabytes of data we cannot move the data where the computing is–instead we must bring the computing to the data. JHU has been one of the pioneers in recognizing this trend and designing systems around this principle [47]. The typical analysis pipeline of a data-intensive scientific problem starts with a low level data access pattern during which outliers are filtered out, aggregates are collected, or a subset of the data is selected based on custom criteria. The more CPU-intensive parts of the analysis happen during subsequent passes. Such analyses are currently implemented in academic Beowulf clusters that combine computeintensive but storage-poor servers with network attached storage. These clusters can handle problems of a few tens of terabytes, but they do not scale above hundred terabytes, constrained by the very high costs of PB-scale enterprise storage systems. Furthermore, as we grow these traditional systems to meet our data needs, we are hitting a “power wall” [48], where the power and space requirements for these systems exceed what is available to individual PIs and small research groups. Existing supercomputers are not well suited for data intensive computations either; they maximize CPU cycles, but lack IO bandwidth to the mass storage layer. Moreover, most supercomputers lack disk space adequate to store PB-size datasets over multi-month periods. Finally, commercial cloud computing platforms are not the answer, at least today. The data movement and access fees are excessive compared to purchasing physical disks, the IO performance they offer is substantially lower (~20MBps), and the amount of provided disk space is woefully inadequate (e.g. ~10GB per Azure instance). Based on these observations, we posit that there is a vacuum today in data-intensive scientific computations, similar to the one that lead to the development of the BeoWulf cluster: an inexpensive yet efficient template for data intensive computing in academic environments based on commodity components. The proposed Data-Scope aims to fill this gap. The Design Concept We propose to develop the Data-Scope, an instrument optimized for analyzing petabytes of data in an academic setting where cost and performance considerations dominate ease of management and security. The Data-Scope will form a template for other institutions facing similar challenges. The following requirements guide the Data-Scope’s design: (a) (b) (c) (d) (e) Provide at least 5 petabytes of storage, with a safe redundancy built in. Keep the ratio of total system to raw disk costs as low as possible. Provide maximal sequential throughput, approaching the aggregate disk speed Allow streaming data analyses on par with data throughput (i.e., 100s of TFlops) Maintain total power requirements as low as possible. This ordered list maps well onto the wish list of most academic institutions. The tradeoff is in some aspects of fault tolerance, the level of automation in data movement and recovery and a certain complexity in programming convenience since the high stream-processing throughput at a low power is achieved by using GPUs. These tradeoffs, combined with maximal use of state of the art commodity components, will allow us to build a unique system, which can perform large data analysis tasks simply not otherwise possible. The Data-Scope will enable JHU scientists and their collaborators to • • • • Bring their 100TB+ data sets to the instrument, analyze them for several months at phenomenal data rates and take their results ‘home’ Create several long-term, robust and high performance services around data sets in the 10200TB range, and turn them into major public resources Explore new kinds of collaborative research in which even the shared, temporary resources can be in the hundreds of terabytes and kept alive for several months Explore new data-intensive computational and data analysis paradigms enabled by the intersection of several technologies (HPC, Hadoop, GPU) and toolkits like CUDA-SQL and MPI-DB. In the paragraphs that follow we describe the Data-Scope’s hardware and software designs. The Hardware Design The driving goal behind the Data-Scope design is to maximize stream processing throughput over TBsize datasets while using commodity components to keep acquisition and maintenance costs low. Performing the first pass over the data directly on the servers’ PCIe backplane is significantly faster than serving the data from a shared network file server to multiple compute servers. This first pass commonly reduces the data significantly, allowing one to share the results over the network without losing performance. Furthermore, providing substantial GPU capabilities on the same server enables us to avoid moving too much data across the network as it would be done if the GPUs were in a separate cluster. Since the Data-Scope’s aim is providing large amounts of cheap and fast storage, its design must begin with the choice of hard disks. There are no disks that satisfy all three criteria. In order to balance these three requirements we decided to divide the instrument into two layers: performance and storage. Each layer satisfies two of the criteria, while compromising on the third. Performance Servers will have high speed and inexpensive SATA drives, but compromise on capacity: Samsung Spinpoint HD103SJ 1TB, 150MB/s, (see [49], verified by our own measurements). The Storage Servers will have larger yet cheaper SATA disks but with lower throughput: Samsung Spinpoint HD203WI 2TB, 110MB/s. The storage layer has 1.5x more disk space to allow for data staging and replication to and from the performance layer. The rest of the design focuses on maintaining the advantages from these two choices. In the performance layer we will ensure that the achievable aggregate data throughput remains close to the theoretical maximum, which is equal to the aggregate sequential IO speed of all the disks. As said before, we achieve this level of performance by transferring data from the disks over the servers’ local PCIe interconnects rather than slower network connections. Furthermore, each disk is connected to a separate controller port and we use only 8-port controllers to avoid saturating the controller. We will use the new LSI 9200-series disk controllers, which provide 6Gbps SATA ports and a very high throughput (we have measured the saturation throughput of the LS92111-8i to be 1346 MB/s). Each performance server will also have four high-speed solid-state disks (OCZ-Vertex2 120GB, 250MB/s read, 190MB/s write) to be used as an intermediate storage tier for temporary storage and caching for random access patterns [48]. Motherboard Memory CPU Enclosure Disk ctrl ext Disk ctrl int Hard disk SSD NIC 10GbE Cables GPU card Performance price Components qty SM X8DAH+F$469 1 18GB $621 1 Intel E5630 $600 2 SM SC846A $1,200 1 N/A LSI 9211-8i $233 3 Samsung 1TB $65 24 OCZ V2 $300 4 Chelsio N310E $459 1 $100 1 2 GTX480 $500 Total Price total $469 $621 $1,200 $1,200 $699 $1,560 $1,200 $459 $100 $1,000 $8,508 Components X8DAH+F-O 24GB Intel E5630 SM SC847 LSI 9200-8e LSI 9211-8i Samsung Chelsio Storage price qty $469 1 $828 1 $600 2 $2,000 3 $338 2 $233 1 $100 126 $540 $100 N/A Total Price 1 3 0 total $469 $828 $1,200 $6,000 $676 $233 $12,600 $540 $300 $0 $22,846 Table 2. The projected cost and configuration for a single unit of each server type The performance server will use a SuperMicro SC846A chassis, with 24 hot-swap disk bays, four internal SSDs, and two GTX480 Fermi-based NVIDIA graphics cards, with 500 GPU cores each, offering an excellent price-performance for floating point operations at an estimated 3 teraflops per card. The Fermibased TESLA 2050 has not been announced yet, we will reconsider if it provides a better price performance as the project begins. We have built a prototype system according to these specs and it performs as expected. In the storage layer we maximize capacity while keeping acquisition costs low. To do so we amortize the motherboard and disk controllers among as many disks as possible, using backplanes with SATA expanders while still retaining enough disk bandwidth per server for efficient data replication and recovery tasks. We will use locally attached disks, thus keeping both performance and costs reasonable. All disks are hot-swappable, making replacements simple. A storage node will consist of 3 SuperMicro SC847 chassis, one holding the motherboard and 36 disks, with the other two holding 45 disks each, for a total of 126 drives with a total storage capacity of 252TB. On Figure 1. The network diagram of the Data-Scope. the storage servers we will use one LSI-9211-8i controller to drive the backplane of the 36 disks, connecting 2x4 SATA ports to the 36 drives, through the backplane’s port multiplier. The two external disk boxes are connected to a pair of LSI 9200-8e controllers, with 2x4 ports each, but the cards and boxes are cross-wired (one 4-port cable from each card to each box), for redundancy in case of a controller failure, as the split backplanes automatically revert to the good controller. The IO will be limited by the saturation point of the controllers and the backplanes, estimated to be approximately 3.6GB/s. Both servers use the same dual socket SuperMicro IPMI motherboard (X8DAH+FO) with 7 PCIeGen2 slots. The CPU is the cheapest 4-core Westmere, but we will be able to upgrade to faster dual 6-cores in the future, as prices drop. In our prototype we tried to saturate this motherboard: we exceeded a sequential throughput of 5GB/s with no saturation seen. servers rack units capacity price power GPU seq IO netwk bw 1P 1.0 4.0 24.0 8.5 1.0 6.0 4.6 10.0 1S 1.0 12.0 252.0 22.8 1.9 0.0 3.8 20.0 90P 90 360 2160 766 94 540 414 900 12S 12 144 3024 274 23 0 45 240 Full 102 504 5184 1040 116 540 459 1140 TB $K kW TF GBps Gbps The network interconnect is 10GbE. Three 7148S switches from Arista Networks’ are Table 3. Summary of the Data-Scope properties for single used at the ‘Top of the Rack’ (TOR), and a servers and for the whole system consisting of Performance high performance 7148SX switch is used (P) and Storage (S) servers. for the ‘core’ and the storage servers. The TOR switches each have four links aggregated to the core for a 40Gbps throughput. We deploy Chelsio NICs, single port on the performance servers and dual port on the storage side. Hardware Capabilities The Data-Scope will consist of 90 performance and 12 storage servers. Table 3 shows the aggregate properties of the full instrument. The total disk capacity will exceed 5PB, with 3PB in the storage and 2.2PB in the performance layer. The peak aggregate sequential IO performance is projected to be 459GB/s, and the peak GPU floating point performance will be 540TF. This compares rather favorably with other HPC systems. For example, the Oak Ridge Jaguar system, the world’s fastest scientific computer, has 240GB/s peak IO on its 5PB Spider file system [50]. The total power consumption is only 116kW, a fraction of typical HPC systems, and a factor of 3 better per PB of storage capacity than its predecessor, the GrayWulf. The total cost of the parts is $1.04M. The projected cost of the assembly + racks is $46K, and the whole networking setup is $114K, for a total projected hardware cost of about $1.2M. In the budget we reserved $200K for contingency and spare parts. The system is expected to fit into 12 racks. Data Ingestion and Recovery Strategies The storage servers are designed for two purposes, Data Replication and Recovery, incremental and full dataset copies and restores (large and small), and Import/Export of Large Datasets, where users show up with a couple of boxes of disks and should be able to start experiments within hours, and keep their data online over the lifetime of the experiment (e.g., months) Individual disk failures at the expected standard rate of about 3%/yr are not expected to cause much of a problem, for the performance servers–this amounts to one failure every 6 days. On our 1PB GrayWulf server we experienced a much lower disk failure rate (~1%) so far. These can be dealt with fairly easily, by reformatting simple media errors automatically with data recovery, and manually replacing failed disks. The bigger challenge is that most of the time the storage servers do not need much bandwidth (e.g., during incremental copies), but there is occasionally a need for considerably more bandwidth for a large restore. Our solution is to design the network for the routine scenarios (i.e., incremental backups and small restores). Both the performance servers as well as the storage servers are configured with hotswappable disks so atypical large restores can be performed by physically connecting disks to the servers (i.e., sneakernet [51]). Given that moveable media (disks) are improving faster than networks, sneakernet will inevitably become the low cost solution for large ad hoc restores, e.g., 10-1000TBs. The hot-swap disks are also useful for importing and exporting large datasets (~100TBs). The DataScope is intended to encourage users to visit the facility and bring their own data. For practical reasons, the data set should be small enough to fit in a few 50 pound boxes (~100TBs). With the hot swappable feature, users could plug in their disks and have their data copied to the performance servers in a few hours. When visitors leave after a few weeks/months, their data could be swapped out and stored in a bookshelf, where it could be easily swapped back in, if the visitor needs to perform a follow-up experiment remotely. Both the performance servers and especially the storage servers could be configured with a few spare disk slots so one can swap in his data without having to swap out someone else’s data. Remote users can transfer data using the fast Open Cloud Consortium (OCC) network [10] – currently a dedicated 10GbE to MAX and Chicago and soon much higher. OCC has also dedicated network links to several Internet NAPs. Finally, the JHU internal backbone is already running at 10Gbps and in the next few months the high throughput genomics facilities at JHMI will be connected to this network. Usage Scenarios We envisage about 20-25 simultaneous applications, which can use the Data-Scope in four different ways. One can run stable, high availability public web services, allowing remote users to perform processing operations on long-lived data sets. These would be typically built on several tens of TB of data and would store data in a redundant fashion for both safety and performance. Examples of such services might be the VO cross-match services in astronomy, or the JHU Turbulence database services. Other applications can load their data into a set of large distributed shared databases, with aggregate sizes in tens to a few hundred TB. The users can run data intensive batch queries against these data sets and store the intermediate and final results in a shared database and file system space. We have developed a parallel workflow system developed for database ingest of data sets in the 100TB range for the PanSTARRS project[52]. This can be turned into a more generic utility with a moderate amount of work. Hadoop is an open source implementation of Google’s MapReduce [53], which provides a good load balancing and an elegant data-parallel programming paradigm. Part of the instrument will run Hadoop over a multitude of data sets. We will experiment with running the most compute-intensive processing stages (bioinformatics, ray-tracing for image simulations in astronomy) on the GPUs using CUDA code. Finally, when all else is inadequate, certain users can request access to the “bare metal”, running their own code end-to-end on the performance servers. User Application Toolkits and Interfaces We will provide users with several general-purpose programming toolkits and libraries to maximize application performance. We have already developed some of the key software components. For example, the custom collaborative environment built for SDSS, Casjobs[54,55], has been in use for seven years, by more than 2,500 scientists. The component we need to add is a shared 100TB intermediate term storage, which has been designed but remains to be built. We have designed and implemented a generic SQL/CUDA interface that enables users to write their own user defined functions that can execute inside the GPUs, but are called from the database. Since all the data flow is on the backplane of the same server, one can achieve a stunning performance. This was demonstrated in our entry for the SC-09 Data Challenge[56]. We have implemented our own S3/Dropbox lookalike, which has been connected to various open source S3 bindings downloaded from SourceForge. This interface is simple, scalable, well-documented, and will provide a convenient way for users to up- and download their smaller data sets. On the applications side we have already ported several key applications to CUDA, but most of the development work will materialize as users start to migrate their applications to the Data-Scope. The expectations are that we will need to customize them for the Data-Scope and integrate them to each other. Other components, such as the integration between MPI and the SQL DB, have been prototyped but will need to be fully developed. In summary, the components that we have developed allow novel high-throughput data analyses. For example, users can, using the SQL-CUDA integration, access powerful analysis patterns like FFT, within a database query. Likewise, Linux MPI applications can read/write data from/to databases using the MPI- DB API. During the initial stages of the Data-Scope’s development we will use some of the performance and storage servers for software development and testing. Data Lifecycles We envisage three different lifecycle types for data in the instrument. The first would be persistent data, over which permanent public services will be built for a wide community, like OpenSkyQuery, the Turbulence database or the Milky Way Laboratory. The main reason to use the Data-Scope in this case is the massive performance gains from the speed of the hardware and parallelism in execution. These data sets will range from several tens to possibly a few hundred TB. The second type of processing will enable truly massive data processing pipelines that require both high bandwidth and fast floating point operations. These pipelines will process hundreds of TB, including reprocessing large images from high throughput genomic sequencers, for reduced error rates, and massive image processing tasks for astronomy or cross correlations of large environmental data sets. Data will be copied physically by attaching 2TB disks to the Data-Scope, while results will be extracted using the same method. These datasets will be active on the system for one to a few weeks. Another typical user of this pattern would be the LHC data analysis group. The third type of usage will be community analysis of very large data sets. Such datasets will be in the 200-500TB range. We will keep the media after the dataset has been copied into the instrument and use them to restore the input data in the case of a disk failure. Once such a massive data set arrives, its partitioning and indexing will be massive endeavor therefore it makes only sense if the data stays active for an extended period (3-12 months). Intermediate, derived data sets could also reach tens or even 100TB. Examples of such datasets include a massive set of simulations (500 cosmological simulations with a high temporal resolution) coupled with an analysis campaign by a broad community. System Monitoring, Data Locality There will be a data locality server, monitoring the file systems, so the system is aware what’s where without depending on a user to update tables manually when disks are swapped in and out. There will be a special file on every disk that tells the system what is on the disk, and a bar code on the outside of disks that would make it easy to locate disks. As for software, we plan to use open source backup software (such as Amanda), as much as possible. Both the operating system software and applications (including Hadoop, SQL Server etc) will be deployed using a fully automated environment, already in use for several years at JHU. Since the performance servers are identical, it is very easy to dynamically change the node allocations between the different usage scenarios. Development Strategy and Methods Our Track Record The JHU group has been systematically working on building high performance data-intensive computing systems (both hardware and software) over the last decade, originally started with Jim Gray. We also have more than a decade of experience in building and operating complex scientific data centers. The 100TB Sloan Digital Sky Survey (SDSS) archive is arguably one of the most used astronomy facilities in the world. Some of the most used web services (OpenSkyQuery) for the Virtual Observatory have also been built and ran out of JHU. Our group has also been hosting the GalaxyZoo project[57] in its first two years until we moved it to the Amazon Cloud. In 2008 we built the 1PB GrayWulf facility (named after our friend Jim Gray), the winner of the Data Challenge at SuperComputing-08, supported by Microsoft and the Gordon and Betty Moore Foundation. The experience gained during the GrayWulf construction was extremely useful in the Data-Scope design. We will also heavily rely on software developed for the GrayWulf[52], but will also add Hadoop to the software provided. It is also clear that the GrayWulf will be maxed out by early Fall 2010, and we need an expansion with ‘greener’ properties, to avoid the power wall. In a project funded by NSF’s HECURA program, we have built low-power experimental systems for data intensive computing, using solid state disks[48]. The lessons learned have been incorporated into the Data-Scope design. JHU was part of the Open Cloud Consortium team[10] winning the Network Challenge at SC-08. Prototypes, Metrics Over the years we have built and customized a suite of performance tools designed to measure and project server performance in data-intensive workloads. We also have access to many years of real workload logs from the SDSS and NVO servers for comparisons. For the Data-Scope proposal we have already built a prototype performance server, which has exceeded the original expectations. The numbers quoted in the proposal are on the conservative side. (d) Impact on the Research and Training Infrastructure Impact on Research There is already a critical mass of researchers at JHU that will use the Data-Scope (see Section (b)). These research problems represent some of the grand challenges in scientific fields ranging from astrophysics and physical sciences, to genome and cancer research, and climate science. What these challenges have in common is that their space and processing requirements surpass the capabilities of the computing infrastructure that the university owns or can lease (i.e., cloud computing). While it is becoming increasingly feasible to amass PB-scale datasets, it is still difficult to share such datasets over wide area networks. The Data-Scope will provide scientists with an environment where they can persistently store their large datasets and allow their collaborators to process these data either remotely (through Web Services) or locally (by submitting jobs through the batch processing system). In this way the Data-Scope will organically become a gathering point for the scientific community. Furthermore, by being a shared environment among multiple projects the Data-Scope will promote sharing of code and best practices that is currently hindered by disparate, vertically integrated efforts. We will seed this community centered at the Data-Scope via our collaborators in other academic institutions and national labs (see letters of collaboration). The availability of compelling datasets at the Data-Scope will have secondary benefits. As the public availability of few-TB datasets (e.g., SDSS), helped nurture research in distributed databases, data mining, and visualization, the ability to broadly access and process multi-PB datasets will create a renaissance in data-intensive computational techniques. Impact on Training The analysis of PB-scale datasets lies at the center of the Fourth Paradigm in Science [58]. Likewise, dealing with the data deluge is of paramount importance to the Nation’s security. Therefore, future generations of scientists and engineers must develop the data analysis skills and computational thinking necessary to compete globally and tackle grand challenges in bioinformatics, clean energy, and climate change. By providing the ability to store, analyze, and share large and compelling datasets from multiple scientific disciplines the Data-Scope will become a focal point in all education levels. The paragraphs that follow describe our interdisciplinary education plans focused around the proposed instrument. The JHU Center for Computational Genomics offers workshops, short courses, a seminar series, and an annual symposium, all of which bring together undergraduate and graduate students, fellows, and faculty (including clinicians) from disciplines as diverse as computer science, molecular biology, oncology, biostatistics, and mathematics. In these courses, participants are encouraged to not only attend courses but also to develop and teach courses to the other “students.” Recent short courses and seminars include discussions of synthetic biology and the database architecture needed for a large-scale synthetic genome project; introduction to the R programming language; biological sequence alignment algorithms; overview of experimental and analytical methods in high-throughput biology, and more. Having such a wide range of expertise and academic experience brings new opportunities for students and professionals in diverse fields and at all levels of training to get involved as well as better communication among scientists when they are part of multi-disciplinary teams. Senior Personnel S. Wheelan will integrate the Data-Scope into future iterations of the workshops offered by the JHU Center for Computational Genomics. Along the same lines, CoPI Church will leverage the NSF-Funded Center for Language and Speech Processing (CLSP) Summer Workshop and the Summer School in Human Language Technologies (HLT), funded by the HLT Center of Excellence, to introduce the Data-Scope to the students in the NLP field. Since its inception in 1998, about 100 graduate students and 60 undergraduates have participated in the workshop. More than 50 additional students supported by the North American Chapter of the Association for Computational Linguistics, and over 20 students from local colleges and universities have attended the two-week intensive summer school portion, leading to the education and training of over 230 young researchers. The workshop provides extended and substantive intellectual interactions that have led indirectly to many continuing relationships. The opportunity to collaborate so closely with top international scientists, both one-on-one and as a team, offers a truly exceptional and probably unprecedented research education environment. Finally, multiple JHU faculty members participating in this project are part of an NSF-funded team on “Modeling Complex Systems: The Scientific Basis of Coupling Multi-Physics Models at Different Scales.” We will leverage this IGERT program to introduce graduate students to the topics of large-scale simulations and data-intensive science using the Data-Scope. Outreach to under-represented communities. CoPI Meneveau will use his existing network to continue recruiting talented Hispanic US graduate students through his contact with Puerto Rico universities and ongoing collaborations with Prof. L. Castillo of RPI and Universidad del Turabo (PR). Over the past 3 years, there have been 3 visiting Hispanic PhD students, 1 MS visiting student from UPR Mayaguez, and 4 REU undergraduate students from Puerto Rico working with Dr. Meneveau. Women are better represented in computational linguistics, environmental sciences, and biology than in computer science and the physical sciences. We will capitalize on this observation to recruit more female students in the sciences that face this gender imbalance. Half of the undergraduate participants in the previously mentioned workshops are women. They are talented students selected through a highly selective search. These students are already involved in inter-disciplinary research in data-intensive science that makes them exceptionally well qualified to enter our PhD programs. We will also leverage the JHU Women in Science and Engineering program (WISE, funded by NSF, housed in the Whiting School of Engineering, and currently linked to Baltimore County’s Garrison Forest School) to build into our program a permanent flow of new students at both the high school and undergraduate levels. The greater Baltimore area has several historically black colleges and universities from which we will actively attempt to recruit students, both at the undergraduate level (for summer research) and as prospective applicants to our program. Our target colleges will include Morgan State University, Coppin State College, Bowie State University, Howard University, and the University of D.C. General outreach. PI Szalay was a member of the NSF Taskforce on Cyberlearning, and is a coauthor of the report “Fostering Learning in the Networked World: The Cyberlearning Opportunity and Challenge”. PI Szalay also led the effort of building a major outreach program around the tens of terabytes of SDSS data. This program has delivered more than 50,000 hours classroom education in astronomy and data analysis for high school students, through a set of student laboratory exercises appropriate for students from elementary through high school. The sdss.org website recently won the prestigious SPORE Award for the best educational websites in science, issued by AAS and Science magazine. Recently the GalaxyZoo (GZ), served from JHU, became one of the most successful examples of `citizen science’ [57]. In this, members of the public were asked to visually classify images of a million galaxies from the SDSS data. More than 100,000 people signed up, did the training course, and performed over 40 million galaxy classifications. GalaxyZoo has been featured by every major news organization in the world (CNN, BBC, the Times of London, NYT, Nature, The Economist, Science News), as an example how science can attract a large involved non-expert population, if presented in the right manner. As part of the NSF-funded projects in Meneveau’s laboratory, two Baltimore Polytechnic Institute (BPI) high school students conducted research activities in his lab. “Baltimore Poly”, as it is affectionately called in this area, has a long tradition of excellence in science and serves a majority African-American student body. CoPI Terzis has collaborated with BPI teachers to develop projects that encourage K-12 students to pursue science and engineering careers through interactive participation in cutting-edge research. This proposed NSF project with the novel capabilities offered by the Data-Scope will enable continuation of the link to Poly’s Ingenuity Project in Baltimore. (e) Management Plan The Work Breakdown Structure (WBS) for the project is found in Table 4. The actual WBS has been developed down to the third level, but has been partially collapsed (bold), in order to fit on a single page. The instrument development will be led by the group of five PIs, also selected as representatives of the different schools of JHU. They will be responsible for the high level management of the project. The management and operations of the instrument (including the sustained operations) will be carried out within the Institute of Data Intensive Engineering and Science (IDIES) at JHU. The system design and architecture is developed by three of the PIs, Alex Szalay, Andreas Terzis and Ken Church, and Jan Vandenberg, Head of Computer Operations at the Dept. of Physics and Astronomy. There is full time Project Manager, Alainna White, who has already been involved in the planning of the schedule and the WBS. She will lead the construction of the instrument hardware and supervise the system software. The design and implementation of the system software will be led by Richard Wilton (with 20 years of experience in SW development), advised by Prof. Randal Burns, from JHU CS. PIs Design/Architecture Project Manager Construction System Software Operations Council Apps Software Operations Figure 2. The organization of the Data-Scope development and operations team. There will be an Operations Council, consisting of 5 faculty members representing different disciplines and a member representing the external users of the instrument. This group will make recommendations during the design, construction and commissioning of the instrument, and make decisions about resource allocation and application support development after the commissioning period. The Operations Council will be chaired by Randal Burns. Additional members include Mark Robbins, Charles Meneveau, Sarah Wheelan, Darryn Waugh and Robert Hanisch (STScI, Director of the VAO, external representative). Operations will be lead by Ani Thakar, who has been responsible for the development and operation of the SDSS Archive, now hosted by JHU who also coordinated the development of the Pan-STARRS data management system. The operations team will consist of a full time System Administrator and a half-time Database Administrator (DBA), shared with IDIES. Alainna White will move into the position of the System Administrator after the commissioning and will also have access to additional support personnel through IDIES in high pressure situations. Extrapolating from our current computing facilities this is adequate support in an academic environment. The parts list and the detailed system configuration has been presented in Section (c), Table 2, since these are closely tied to the goals and performance of the instrument. The total cost of the hardware parts is $1,040K, networking $114K, racks and assembly $46K. There is a $200K contingency, to accommodate price fluctuations, and spare parts. A large fraction of this will be funded by JHU’s cost sharing ($980K). The first month of the project is spent on Hardware Configuration (Task 1), reevaluating the original, existing design, accommodate possible new emerging hardware components with a better price performance, update the in-house prototypes and reconfirm the performance metrics. There is minimal risk in this phase, since we have already built and validated the performance server, and the storage server is very straightforward in all respects. The only possible risk is a sudden escalation of CPU or memory prices, in which case we cut back on purchasing the full memory. We also plan to delay the purchase of half the GPU cards, since this is the field with the fastest pace of evolution, every three months there are new, better and cheaper cards on the market. With the recent release of the first NVIDIA Fermi, this is expected to consolidate by the end of 2010. The System Software (Task 2) is a main part of the system. Most of the important pieces have been largely developed or designed for the SDSS, GrayWulf, Pan-STARRS and the turbulence archives. Some have been in use by thousands of people for more than 6 years (Casjobs). This helps to mitigate the risks involved. Nevertheless, there will be still a lot of extra development needed to modify, homogenize and generalize these tools. These modification tasks will be carried out by the same individuals who developed the original software, thus there will be a relatively mild learning curve. Some of this effort can be started right away, on the prototype boxes, while the rest of the system is assembled. The main categories for the system software have not been collapsed, so that they can be seen individually. The total system software development comes to 24 man-months, all during the first year. The Construction schedule (Task 3) is rather aggressive. The main risk is that some of the components may be out of stock (at a good price) given the quantity. We encountered this in the past with SSDs. This is why we budgeted 28 days for the assembly and preparation. We can absorb another two weeks lead time without a major crisis. The preparation of the instrument room and networking will begin immediately as the design is consolidated. We allocated 14 days for validation and testing of the different hardware components. Over the years we have built a lot of automated diagnostic and test tools to validate the performance of the different system components, therefore risks are minimal. Since we will have some spare parts, faulty boxes can be repaired on site. Disks will be directly shipped to JHU, and inserted into the servers after they are rack mounted, to minimize disk damages during shipping. We will hire undergraduates for the disk tray assembly, bar coding and insertion. During the Commissioning phase (Task 4) we will installing user interfaces, create the environments for the test users including the loading a several large data sets. We will also simulate various system failures and recoveries. End user documentation will be prepared and revised during this period. We have budgeted an additional 12 months in year 2 to interface system components with the evolving applications, since it is clear that there will some unanticipated requests made by the different apps that have implications for the system software. After a decade in developing data-intensive computing hardware and software systems we feel that the task at hand (with the 12 months in year 2) can be realistically accomplished, given the reuse of our existing software base, as part of the commissioning phase. Application Support (Task 5) will be a major part of our development, a total of 64 months of effort will be spent on enhancing existing applications to maximally utilize the Data-Scope, like CUDA porting of application kernels, custom data partitioning tools, Hadoop conversion, DB loading workflow customization. There will be dedicated SW engineers, whose development time will be allocated in approximately 3 month chunks by the Operations Council and supervised by the Head of Operations. In the Operations Phase (Task 6, not shown due to space constraints), starting in Jan 2012, we reevaluate every six months (by the Operations Council) how the resources will be allocated among the different projects, and the lifecycles of the different data sets. The reallocation may include moving some of the machines from a Windows DB role to Linux – easily done since the HW is identical, and SW install is automatic push. We expect requests to come in the form of a short (1 page) proposal. In exceptional cases the 6 month cycle can be accelerated. Users can either use networks to copy their data sets, or as described in Section(c), they can bring their own disks, still the most efficient way to move large data. JHU is committed to a 5 year Sustained Operation of the Data-Scope beyond the end of the MRI grant (see support letter). In this phase, JHU will provide the facility, electricity and cooling, the salary of the System Manager, the 50% DBA, and the MAX connection fees. As hardware upgrades become necessary, we envisage first upgrading the disks (as new projects bring their own disks, they will leave them there). CPU and memory upgrades are incremental and easy, and they will be financed by projects needing the enhanced capabilities. We expect that the 5PB capability of the system will be maxed-out in about 2-3 years. At that point we hope that the system becomes so indispensible to many of the involved research projects that they will use their own funds to incrementally add to the system. In a ‘condominium model’, that IDIES has been using to sustain HPC at JHU, users can buy blocks of new machines, which gives them a guaranteed access to a time-share of the whole instrument. The configuration of these blocks is standardized, to benefit from the economy of scale in automated system management. All software developed is and will be Open Source. Our GrayWulf system has been cloned at several places, the Casjobs environment is running at more than 10 locations world-wide. Our data access and web services applications (and our data sets) have been in use over 20 institutions world-wide. We will create a detailed white paper on the hardware design, the performance metrics, and create scaled-down reference designs containing concrete recipes in the Beowulf spirit, for installations smaller than the DataScope, to be used by other institutions. WBS 1 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 2 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 2.10 2.11 2.12 2.13 2.14 3 3.1 3.2 3.3 3.4 3.5 4 4.1 4.2 4.3 4.4 4.5 4.6 5 5.1 5.2 5.3 5.4 5.5 Task Name Hardware Configuration Reevaluate components (price and performance) Reevaluate detailed design Update prototype performance box Update prototype storage box Update and Verify Performance Metrics Update Network architecture Update requirements for operating environment Design detailed recovery plan System Software Development Create a data map application/service Create MPI-DB Create SQL-CUDA Framework Create recovery software Customize system monitoring software Customize sensor monitoring software Create S3/Dropbox service Create customized interface for Dropbox Customize MyDB / CASJobs Create monitoring database application and interface Loading Framework Disk Tracking Software Design automated OS/SW installation environment Create deployment plan Construction Select vendor for off-site assembly Order all components Assembly and Preparation System Integration Initial system testing and debug Commissioning Establish WAN Connections Ingest of Datasets Install User Interaction Layer Create user test environment Simulate failures and recoveries Unanticipated software build requests Application Software Support Data partitioning Loading workflow customization Application DB conversion Hadoop Conversion Application kernel porting to CUDA WBS Code (1) HW Configuration (2) Sys SW Dev (3) Construction (4) Commissioning (5) App SW Support 1/11 2/11 3/11 4/11 5/11 6/11 7/11 Duration 26 days 10 days 10 days 10 days 10 days 5 days 5 days 3 days 7 days 180 days 60 days 180 days 27 days 60 days 85 days 60 days 60 days 35 days 90 days 60 days 60 days 30 days 20 days 8 days 84 days 10 days 14 days 28 days 18 days 14 days 412 days 12 days 60 days 30 days 150 days 60 days 260 days 475 days 475 days 475 days 475 days 475 days 475 days 8/11 9/11 Start 1/3/11 1/3/11 1/12/11 1/17/11 1/17/11 1/31/11 1/10/11 2/3/11 1/17/11 1/3/11 1/3/11 1/3/11 4/8/11 1/3/11 1/11/11 1/3/11 5/2/11 7/25/11 1/3/11 1/11/11 1/3/11 1/3/11 1/28/11 1/24/11 2/7/11 2/7/11 2/21/11 3/11/11 4/20/11 5/16/11 6/3/11 6/3/11 6/3/11 6/3/11 6/3/11 6/3/11 1/3/12 3/1/11 3/1/11 3/1/11 3/1/11 3/1/11 3/1/11 10/11 11/11 Finish 2/7/11 1/14/11 1/25/11 1/28/11 1/28/11 2/4/11 1/14/11 2/7/11 1/25/11 9/9/11 3/25/11 9/9/11 5/16/11 3/25/11 5/9/11 3/25/11 7/22/11 9/9/11 5/6/11 4/4/11 3/25/11 2/11/11 2/24/11 2/2/11 6/2/11 2/18/11 3/10/11 4/19/11 5/13/11 6/2/11 12/31/12 6/20/11 8/25/11 7/14/11 12/29/11 8/25/11 12/31/12 12/24/12 12/24/12 12/24/12 12/24/12 12/24/12 12/24/12 12/11 2012 Existing Facilities, Equipment and Other Resources Our collaboration has a substantial amount of existing hardware and facilities, which will enhance the possible uses of the Data-Scope. These facilities have also served as a way to gradually understand the real needs of data-intensive computing, and as prototypes for our current, much larger instrument. The DLMS Cluster The DLMS cluster was supported by an NSF MRI grant, MRI-320907 DLMS: Acquisition of Instrumentation for a Digital Laboratory for Multi-Scale Science, PIs: Chen et al.). The cluster consists of a compute layer and a database layer, the JHU public turbulence database. It houses a 27 TB database that contains the entire time history of a 10243 mesh point pseudo-spectral DNS of forced isotropic turbulence, connected to a 128 node Beowulf cluster througha Myrinet fabric. This was our first experiment in integrating traditional HPC with databases. The resulting public database has been in wide use for many years (see Section (b) of the Project Description), and has recently been migrated to the Graywulf, for improved performance. The GrayWulf Cluster GrayWulf is a distributed database cluster at JHU consisting of 50 database nodes with 22TB and an 8-core server each, for a total of 1.1PB. The cluster was purchased on funds from the Gordon and Betty Moore Foundation, the Pan-STARRS project and Microsoft Research. The cluster already hosts several large data sets (Pan-STARRS, turbulence, SDSS, various Virtual Observatory catalogs and services, environmental sensor data, computer security data sets, network traffic analysis data, etc). Currently about 500TB is already utilized. The cluster has an IO performance exceeding many supercomputers: the aggregate sequential read speed is more than 70 Gbytes/sec. The GrayWulf is a direct predecessor for the Data-Scope. One of its weaknesses is that there is no low-cost storage layer, backups were made to tapes, a soluition that is not scalable in the long run. The HHPC Cluster The same computer room hosts an 1800 core BeoWulf cluster, a computational facility shared among several JHU faculty. The HHPC and the GrayWulf share a common 288-port DDR Infiniband switch for an extremely high-speed interconnect. There is an MPI interface under development that will enable very fast peer-to-peer data transfers between the compute nodes and the database nodes. The Deans of the JHU Schools provide funds to cover the management and operational costs of the two connected clusters as part of IDIES. The HECURA/Amdahl Blade Cluster As part of our ongoing HECURA grant (OCI-09104556) we have built a 36-node cluster combining low power motherboards, GPUs and solid state disks. The cluster, while only consuming 1.2kW, has an IO speed of 18Gbytes/sec, and using the GPU/SQL integration, it was able to compute 6.4 billion tree traversals for an astronomical regression problem. NVIDIA has also provided a substantial support (hardware donation and a research grant) for this system. NSF-MRI NVIDIA cluster JHU has been recently awarded an NSF MRI grant (CMMI-0923018), to purchase a large GPU cluster. We are in the process of architecting and ordering this system based on the next generation Fermi architecture (as soon as the first Fermi-based TESLA cards are available). The cluster will also have a high speed disk IO. An additional significance of the cluster is that it will help to introduce many students to this technology. There is a natural cohesion between the Data-Scope and the GPU cluster. The Open Cloud Consortium Testbed The OCC has placed so far one rack of a BeoWulf cluster, and Yahoo is in the process of deploying two more racks of the cluster to JHU. There will be five more racks (42 nodes each with 8 cores and 3TB) in Chicago, for a total of almost 900TB of disk space. The servers are connected to a 48-port 1Gbps switch, with two 10Gbps uplink ports. This system is part of the Open Science Cloud Testbed, and can be used for the Hadoop-based transform and load workflows distributed over geographic distances. The purpose of the cluster is to explore petascale distributed computing and data mining challenges where the nodes are separated across the continental US. There is a dedicated 10Gbps lambda connection from the JHU cluster to UIC, via the id-Atlantic Crossroads (MAX), and McLean VA. The Infiniband switch of the HHPCGrayWulf has also a 10Gbit module that is connected to the outgoing 10Gbps line. This enables any one of the GrayWulf machines to be accessible from any of the OCC servers. The core switch of the DataScope will be also directly linked to the 10Gbps backbone. JHU Internal Connections The internal JHU backbone has recently been upgraded to 10Gbps. The Data-Scope will be able to connect directly to the 10Gbps router. The different partners within JHU will be all connected to the fast backbone during the first two years of the project. References [1] Sloan Digital Sky Survey Archive, http://skyserver.sdss.org/ [2] Virtual Astronomical Observatory, http://us-vo.org/ [3] Budavari,T., Szalay,A.S, Malik,T., Thakar,A., O'Mullane,W., Williams,R., Gray,J., Mann,R., Yasuda,N.: Open SkyQuery -- VO Compliant Dynamic Federation of Astronomical Archives, Proc. ADASS XIII, ASP Conference Series, eds: F.Ochsenbein, M.Allen and D.Egret, 314, 177 (2004). [4] Large Synoptic Survey Telescope, http://lsst.org/ [5] Panoramic Survey Telesope and Rapid Response System, http://pan-starrs.ifa.hawaii.edu/ [6] Klypin, A., Trujillo-Gomez, S, Primack, J., 2010, arXiv:1002.3660. [7] Diemand, J., Kuhlen, M., Madau, P., Zemp, M., Moore, B., Potter,D., & Stadel, J. 2008, Nature, 454, 735 [8] Heitmann, K., White, M., Wagner, C., Habib, S., Higdon, D., 2008, arXiv:0812.1052v1 [9] Steinmetz, M. et al. 2006, AJ, 132, 1645 [10] Open Cloud Consortium, http://opencloudconsortium.org/ [11] Sarah Loebman, Dylan Nunley, YongChul Kwon, Bill Howe, Magdalena Balazinska, and Jeffrey P. Gardner, 2009, Proc. IASDS. [12] Yeung, P.K., R.D. Moser, M.W. Plesniak, C. Meneveau, S. Elgobashi and C.K. Aidun, Report on NSF Workshop on Cyber-Fluid Dynamics: New Frontiers in Research and Education, 2008. [13] Moser, R.D., K. Schulz, L. Smits & M. Shephard, “A Workshop on the Development of Fluid Mechanics Community Software and Data Resources”, Report on NSF Workshop, in preparation, 2010. [14] Perlman, E. R. Burns, Y. Li & C. Meneveau, Data exploration of turbulence simulations using a database cluster, In Proceedings of the Supercomputing Conference (SC’07), 2007. [15] Li, Y., E. Perlman, M. Wan, Y. Yang, C. Meneveau, R. Burns, S. Chen, G. Eyink & A. Szalay, A public turbulence database and applications to study Lagrangian evolution of velocity increments in turbulence, J. Turbulence 9, N 31, 2008. [16] Chevillard, L. and C. Meneveau, Lagrangian dynamics and statistical geometric structure of turbulence, Phys. Rev. Lett. 97, 174501, 2006. [17] Chevillard, L. and C. Meneveau, Intermittency and universality in a Lagragian model of velocity gradients in three-dimensional turbulence, C.R. Mecanique 335, 187-193, 2007. [18] Li, Y. and C. Meneveau, Origin of non-Gaussian statistics in hydrodynamic turbulence, Phys. Rev. Lett., 95, 164502, 2005. [19] Li, Y. and C. Meneveau, Intermittency trends and Lagrangian evolution of non-gaussian statistics in turbulent flow and scalar transport, J. Fluid Mech., 558, 133-142, 2006. [20] Li, Y., C. Meneveau, G. Eyink and S. Chen, The subgrid-scale modeling of helicity and energy dissipation in helical turbulence, Phys. Rev. E 74, 026310, 2006. [21] Biferale, L., L. Chevillard, C. Meneveau & F. Toschi, Multi-scale model of gradient evolution in turbulent flows, Phys. Rev. Letts. 98, 213401, 2007. [22] Chevillard, L., C. Meneveau, L. Biferale and F. Toschi, Modeling the pressure Hessian and viscous Laplacian in turbulence: comparisons with DNS and implications on velocity gradient dynamics, Phys. Fluids 20, 101504, 2008. [23] Wan, M., S. Chen, C. Meneveau, G. L. Eyink and Z. Xiao, Evidence supporting the turbulent Lagrangian energy cascade, submitted to Phys. Fluids, 2010. [24] Chen, S. Y., G. L. Eyink, Z. Xiao, and M. Wan, Is the Kelvin Theorem valid for highReynolds-number turbulence? Phys. Rev. Lett. 97 144505, 2006. [25] Meneveau, C. Lagrangian dynamics and models of the velocity gradient tensor in turbulent flows. Annu. Rev. Fluid Mech. 43, in press, 2010. [26] Yu H. & C. Meneveau. Lagrangian refined Kolmogorov similarity hypothesis for gradient time evolution and correlation in turbulent flows. Phys. Rev. Lett. 104, 084502, 2010. [27] Eyink, G. L., Locality of turbulent cascades, Physica D 207, 91-116, 2005. [28] Eyink, G. L., Turbulent cascade of circulations, Comptes Rendus Physique 7 (3-4), 449-455, 2006a. [29] Eyink, G. L., Multi-scale gradient expansion of the turbulent stress tensor, J. Fluid Mech. 549 159-190, 2006b. [30] Eyink, G. L. Turbulent diffusion of lines and circulations, Phys. Lett. A 368 486–490, 2007. [31] Eyink, G. L. and H. Aluie, The breakdown of Alfven’s theorem in ideal plasma. flows: Necessary conditions and physical conjectures, Physica D, 223, 82–92, 2006. [32] Eyink, G. L., Turbulent flow in pipes and channels as cross-stream “inverse cascades” of vorticity, Phys. Fluids 20, 125101, 2008. [33] Eyink, G. L., Stochastic line motion and stochastic conservation laws for nonideal hydromagnetic models, J. Math. Phys., 50 083102, 2009. [34] Eyink, G. L. The small-scale turbulent kinematic dynamo, in preparation, 2010. [35] Luethi, B., M. Holzer, & A. Tsinober. Expanding the QR space to three dimensions. J. Fluid Mech. 641, 497-507, 2010. [36] Gungor A.G. & S. Menon. A new two-scale model for large eddy simulation of wall-bounded flows. Progr. Aerospace Sci. 46, 28-45, 2010. [37] Wang, J., L.-P. Wang, Z. Xiao, Y. Shi & S. Chen, A hybrid approach for numerical simulation of isotropic compressible turbulence, J. Comp. Physics, in press, 2010. [38] Mittal, R. Dong H., Bozkurttas, M., Najjar F.M., Vargas, A. and vonLoebbecke A., "A versatile sharp interface immersed boundary method for incompressible flows with complex boundaries", J. Comp. Phys. 227, 4825-4852, 2008. [39] Le Maître, O.P. L. Mathelin, O.M. Knio & M.Y. Hussaini, Asynchronous time integration for Polynomial Chaos expansion of uncertain periodic dynamics, Discrete and Continuous Dynamical Systems 28, 199-226, 2010. [40] Le Maître O.P. & O.M. Knio, Spectral Methods for Uncertainty Quantification – With Application to Computational Fluid Dynamics, Springer, 2010. [41] Szalay,A.S., Gray,J., 2006, Science in an Exponential World, Nature, 440, 413. [42] Bell, G., Gray,J. & Szalay, A.S. 2006, “Petascale Computational Systems: Balanced CyberInfrastructure in a Data-Centric World”, IEEE Computer, 39, pp 110-113. [43] Bell, G., Hey, A., Szalay, A.S. 2009, “Beyond the Data Deluge”, Science, 323, 1297 [44] Moore’s Law: http://en.wikipedia.org/wiki/Moore%27s_law [45] Walter, C., 2005, Kryder’s Law. Scientific American. August 2005. [46] Nielsen’s Law: http://www.useit.com/alertbox/980405.html [47] Szalay,A.S., Gordon Bell, Jan Vandenberg, Alainna Wonders, Randal Burns, Dan Fay, Jim Heasley, Tony Hey, Maria Nieto-SantiSteban, Ani Thakar, Catherine Van Ingen, Richard Wilton: GrayWulf: Scalable Clustered Architecture for Data Intensive Computing, Proceedings of the HICSS-42 Conference, Mini-Track on Data Intensive Computing, ed. Ian Gorton (2009). [48] Szalay, A., Bell, G., Huang, H., Terzis, A., White, A. Low-Power Amdahl-Balanced Blades for Data Intensive Computing. In the Proceedings of the 2nd Workshop on Power Aware Computing and Systems (HotPower '09). October 10, 2009. [49] Schmid, P., Ross, A., 2009, Sequential Transfer Tests of 3.5” hard drives, http://www.tomshardware.com/reviews/wd-4k-sector,2554-6.html [50] Spider: http://www.nccs.gov/computing-resources/jaguar/file-systems/ [51] Church, K., Hamilton, J.,, A. 2009, Sneakernet: Clouds with Mobility, http://idies.jhu.edu/meetings/idies09/KChurch.pdf [52] Yogesh Simmhan, Y., Roger Barga, Catharine van Ingen, Maria Nieto-Santisteban, Laszlo Dobos, Nolan Li, Michael Shipway, Alexander S. Szalay, Sue Werner, Jim Heasley, 2009, GrayWulf: Scalable Software Architecture for Data Intensive Computing, Proceedings of the HICSS-42 Conference, Mini-Track on Data Intensive Computing, ed. Ian Gorton. [53] J. Dean and S. Ghemawat, “MapReduce: Simplified Data Processing on Large Clusters”, 6th Symposium on Operating System Design and Implementation, San Francisco, 2004. [54] W. O’Mullane, N. Li, M.A. Nieto-Santisteban, A. Thakar, A.S. Szalay, J. Gray , 2005, “Batch is back: CasJobs, serving multi-TB data on the Web,”, Microsoft Technical Report, MSR-TR2005-19. [55] Li, N. Szalay, A.S., 2009, “Simple Provenance in Scientific Databases”, Proc. Microsoft eScience Workshop, Pittsburgh, eds: T. Hey and S. Tansley. [56] Szalay,A.S. et al, 2009: Entry in Supercomputing-09 Data Challenge. [57] Raddick J., Lintott C., Bamford S., Land K., Locksmith D., Murray P., Nichol B., Schawinski K., Slosar A., Szalay A., Thomas D., Vandenberg J., Andreescu D., 2008, “Galaxy Zoo: Motivations of Citizen Scientists”, Bulletin of the American Astronomical Society, 40, 240 [58] The Fourth Paradigm, Data-Intensive Scientific Discovery, 2009, eds; T. Hey, S. Tansley, K. Tolle, Microsoft Research Press. MRI Coordinator The National Science Foundation 4201 Wilson Boulevard, Arlington, VA 22230 Charlottesville, April 20, 2010 Dear Sirs, We would like to express NVIDIA’s interest in the proposal entitled “Data-Scope – a Multi-Petabyte Generic Analysis Environment for Science”, submitted to the NSF MRI program by the Johns Hopkins University, PI: Alexander Szalay. We are very excited about the possibility of building a new, balanced architecture for the emerging dataintensive computational challenges. These problems do not map very well unto the traditional architectures. The unique combination of extreme I/O capabilities combined with the right mix of GPUs represent a novel approach we have not seen anywhere else and has a very good chance to result in major breakthroughs in how we think about future extreme scale computing. Furthermore, the integration of CUDA with the SQL Server engine and the ability to run GPU code inside User Defined Functions will give database queries unprecedented performance. NVIDIA is happy to support this effort, and in recognition of his work, JHU became one of the inaugural members of our recently formed CUDA Research Centers. We will provide JHU with advance technical information on technologies and on our emerging new products, send evaluation boards, and we will also extend major discounts as NVIDIA equipment is acquired. We believe that the design of the Data-Scope instrument described in the proposal will find a broad applicability, and we will see it replicated in many universities around the world. Sincerely, David Luebke, Ph.D. NVIDIA Distinguished Inventor Director of Research NVIDIA Corporation