Answer Key for Activity #2
advertisement

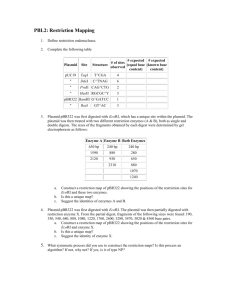
Teacher Notes for Activity 2 Choosing a Plasmid Choosing the proper plasmid for your cloning experiment is very important. There are many options available and being able to choose one that will be optimal for your particular experiment can be daunting! In this activity, you will examine four commonly used Promega plasmids to determine their characteristics and which will be the best choice for your “experiment.” Examine the four plasmids provided. Your options are pBR322, pCAT® 3-Basic, pF1A T7 Flexi® and pGEM® 3Z and answer the following questions. (Note: the plasmids that are included in the student handout are also available as a PowerPoint slide (plasmids.ppt) so that you can project them if you care to discuss them in class. Answers to questions are provided in bold italics below. 1) Which plasmid(s) will grow on plates treated with Ampicillin? All of the plasmid have Amp’ resistance 2) Which plasmid(s) will grow on plates treated with Tetracycline? Only pBR322 has Tet’ resistance 3) If you choose pBR322 as your vector and digest it with Sca l and insert your gene of interest, will it be able to grow in the presence of Ampicillin? Why or why not? It will not grow in the presence of Ampicillin because the resistance gene will have been altered by the insertion of the foreign DNA 4) Which plasmid(s) have a restriction site for Hind lll? pBR322, pCAT® 3-Basic, and pGEM® 3Z 5) Do any of the plasmids have more than one restriction site for the same enzyme? If so, would that be a good enzyme to choose for your digests? Why or why not? Yes, some of the plasmids have multiple restriction sites for the same enzyme (ex. pCAT® 3-Basic has two sites for Hind lll. This would not be a good choice since we only want one opening in our plasmid. Teacher Notes for Activity 2 Choosing a Plasmid Page 1 of 4 Contextual Biology Integrated Projects Created by the Center for Occupational Research and Development http://www.cordonline.net/HiESTbiology Teacher Notes for Activity 2 Choosing a Plasmid Page 2 of 4 Contextual Biology Integrated Projects Created by the Center for Occupational Research and Development http://www.cordonline.net/HiESTbiology Now that you have examined the plasmids, choose which plasmid you would use considering the following parameters: Restriction enzymes can be expensive; therefore, it is often a good idea to use an enzyme that you or colleagues already have in the lab. The same is true for antibiotics. In your lab you have Ampicillin and Tetracycline; you also have the restriction enzymes Bam Hl (G'GATCC) and Eco Rl (G’AATTC). You are going to be inserting a 550 kb fragment of DNA into your chosen vector. The sequence of your fragment is found at the end of this document or in the separate document sequence.doc that your teacher can provide you. (Note: You may want to load the document with the plasmid sequence (sequence.doc) on to the computers students will be using so that they can easily cut and paste the sequence.) Your sequence must be cut with the same enzyme as the plasmid. Use the NEB cutter online tool found at the following url: (http://tools.neb.com/NEBcutter2/index.php) to determine restriction sites within your sequence. In NEB cutter, you will cut and paste your sequence into the pane, select “all commercially available enzymes” and click “submit.” The cutter will show you all the restriction enzyme sites within your sequence. Which plasmid, restriction enzyme and antibiotic should you choose for your experiment? WHY? The NEB cutter data will show that our sequence does not have a restriction site for Bam Hl. It does have one site for EcoRl. Therefore, we will digest with EcoRl. Since our lab routinely uses tetracycline, the plasmid of choice will be pBR322. Since the EcoRl site does not interfere with the Tet’ resistance that will be the best choice for our experiment! Teacher Notes for Activity 2 Choosing a Plasmid Page 3 of 4 Contextual Biology Integrated Projects Created by the Center for Occupational Research and Development http://www.cordonline.net/HiESTbiology Online Sequence 061 121 181 241 301 361 421 481 541 601 661 721 781 841 901 961 1021 1081 1141 1201 tgtcttgttt ggagattagg gagaaagtta tcatttgaca ccatgctgac acaagccgct ctcagggtca ctcacacatg aggggccttc ggagacagca cagggggtag accagaccct caacaacaaa aaaagagggt gagtaaaggg gttcatagct ggagggtctt ttctttagag gagaggggag gttcttgtgt aaatttttgg agggaggaag gacaagtaga gtcactctgg agtgcaccat gctcgtgggc ctgtgtttag atcacattat atatgccttc gtccagcaac tgcagaggct aggcaggtga aaaattagag tatgatttgg aatttttctt gggcatggta tggaggccag agagaggaga aggagagagg gttcattggt Teacher Notes for Activity 2 Choosing a Plasmid Page 4 of 4 aaacaagtat taaacaaggg tgagatggtt ctccctgacg atatgcatat acctgtctca tgtcgctggg catgtgacta tctaacatag cactcctaga taggagatag ctgaccttgt acagatatcc aatgcactac ggcaaaaggg gctcatacgt gagtttgaga gaggggagag gcagaagaga ggagatgctg atattacttt aggagaggag atagggacga aatcaaaaca gtaacaggga gctctagcct ttatacactg gcacctacta tcagagggta gaggaaggag agccaggctt ctgaatctca ccaaatatgt tgtaaaccac agaagttcac gtaatcacag ccagcctggg gggagaggag attccttggt ttcctgggca catagttgaa aaaggcagga atgaagctag gccacccagg acagcagccc cagctcaggc gacaagtacc gagccgcaca aggtaataag ttcaggccag gaatctgctg tatgtaaaat caaatttatt aggcacgtgc ataagctgct ctatgtggga caatgtgatg agagaagaca tctgaaggct ggtgttcttt aagaaagtga agaggagaga aattgtgaaa cagtccttgt cggctggagc ttcacttagc agtggcacca tcctgtctgg cacagggagg caaacaaggg ctacctgact gcagtgtcaa caggaataag agtgagggaa tagaaacaga ggctgaggcg aggctccagt ggggacgggg caaagccaga tgacagcatc Contextual Biology Integrated Projects Created by the Center for Occupational Research and Development http://www.cordonline.net/HiESTbiology