Polygenic inheritance
advertisement

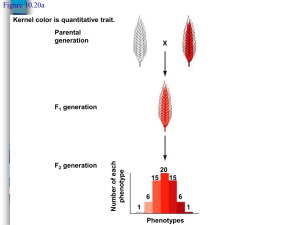
IB Biology HL Genetics - Polygenic inheritance. One of the major problems in genetics during the early part of the 20th century involved the following question. If Mendel's ideas were correct then how can one explain the inheritance of quantitative traits where the offspring of a cross tended to be intermediate in appearance between the two parents. For instance if one parent is tall and the other short, the offspring tend to be intermediate in height. In other words, the offspring in a cross tend to be a blend of both parents. Inheritance like this is called polygenic inheritance. Assumptions of the Polygenic Model: This model makes the following 6 simplifying assumptions: Each contributing gene has small and relatively equal effects. The effects of each allele are additive. The genes at each locus behave as if they follow Codominance. There is no linkage involved. The value of the trait depends solely on genetics; environmental influences can be ignored . Example; Polygenic inheritance of color in wheat. Kernal color in wheat is determined by two gene pairs that produce a range of colors from white to dark red depending on the combinations of alleles. Dark red plants are homozygous AABB and white plants are homozygous aabb. When these homozygotes are crossed the F1 offspring are all double heterozygotes AaBb. Thus crossing individuals with the phenotype extremes yield offspring that are a 'blend' of the two parents. This illustrates an important point that many times when you have two parents who differ in phenotype for some characteristic, there is a tendency for the offspring to be intermediate to the parents in phenotype. But what happens when the two double heterozygotes are crossed? The results are shown in the following Punnett Square Notice that there are 5 phenotypes - corresponding to the number of upper case alleles 0 through 4 that can be present in the offspring. Observe too that even though both parents are intermediate, there is not blending in the offspring. Offspring that can be more extreme than either parent- 1/16th of the offspring are dark red and 1/16 are white. Even though the polygenic model makes a number of simplifying assumptions it does seem to be a good approximation to the inheritance of a large number of quantitative traits. AB Ab aB ab AB AABB AABb AaBB AaBb Ab AABb AAbb AaBb Aabb aB AaBB AaBb aaBB aaBb ab AaBb Aabb aaBb aabb Questions 1. Which allele A or a is dominant? 2. Which Gene A or B is dominant? 3. Put these genotypes in order of darkness: AAbb , AaBB , AABB , aaBb. 4. How many different colours (Phenotypes) are there? 5. Sketch a bar chart to show the 1: 4: 6: 4 :1 ratio of phenotypes 6. Now complete section 10.3.2 in the revision booklet….. Polygenic inheritance of plant height in tobacco: Plant height in tobacco is controlled by a series of genes at multiple loci that each has a small additive affect on the phenotype of the plant. Assume three loci, each of which has two alleles. (A,a B,b C,c). Imagine pure-breeding short plants are all aabbcc and tall plants are all AABCC and a situation where the height of the plant is determined entirely by the number of upper case alleles regardless of which locus the allele is at. Thus a plant with the genotype AaBbcc is the same height as a plant with genotype AabbCc. In contrast to the claim of your text the upper case alleles are not dominant but behave as incomplete dominant alleles. There are 7 possible classes of plant heights depending on the number of upper case alleles. 0,1,2,3,4,5 or 6. Consider a pure breeding short plant aabbcc crossed with a pure breeding AABBCC plant. The F1's resulting from this cross are clearly the triple heterozygote: AaBbCc Notice that these plants are going to be intermediate in height between the two parents. But what happens when these intermediate individuals are bred with each other? To analyze this, assume that the gene pairs are unlinked. This allows us to use independent assortment to predict the results. The expected fraction of offspring in each height class is given by the binomial theorem: Notice that the frequency distribution of phenotypes in the F2 generation looks a little like the bell shaped curve familiar to students of statistics as the 'Normal Distribution'. Indeeed for large numbers of genes invovled in a quantitative trait where each gene has a small additive effect the resulting distribution of phenotype classes very closely resembles the Normal Distribution. Polygenic Trait: Some traits, some phenotypes, are controlled by more than one gene. It was mentioned in the monohybrid cross, above, that technically, human eye color is controlled by at least two genes, one which codes for brown vs. blue and another which codes for green vs. blue. In the epistasis crosses, below, you will see other examples of polygenic traits. Human skin color is also a classic example of a polygenic trait. It is known that at least three or four genes control skin color, and for each of those genes, dark pigment has incomplete dominance over light (so a heterozygote would be intermediate — see above). Because we just did a trihybrid cross, let’s assume three genes here (for simplicity), and to avoid confusion among them, let’s arbitrarily call them genes A, B, and C. Then, someone who is AABBCC would have very dark skin color and someone who is aabbcc would have very light skin color. If they would get married and have children, their children would all be AaBbCc. If two of those people would get married and have children, the Punnett square would look like this: ABC ABc AbC Abc aBC aBc abC abc ABC AABBCC AABBCc AABbCC AABbCc AaBBCC AaBBCc AaBbCC AaBbCc ABc AABBCc AABBcc AABbCc AABbcc AaBBCc AaBBcc AaBbCc AaBbcc AbC AABbCC AABbCc AAbbCC AAbbCc AaBbCC AaBbCc AabbCC AabbCc Abc AABbCc AABbcc AAbbCc AAbbcc AaBbCc AaBbcc AabbCc Aabbcc aBC AaBBCC AaBBCc AaBbCC AaBbCc aaBBCC aaBBCc aaBbCC aaBbCc aBc AaBBCc AaBBcc AaBbCc AaBbcc aaBBCc aaBBcc aaBbCc aaBbcc abC AaBbCC AaBbCc AabbCC AabbCc aaBbCC aaBbCc aabbCC aabbCc abc AaBbCc AaBbcc AabbCc Aabbcc aaBbCc aaBbcc aabbCc aabbcc So, to summarize, as above: AABBCC (6 dark) AABBCc (5 dark) 1 2 genotypes: AaBBCC (5 dark) AaBBCc (4 dark) 2 4 aaBBCC (4 dark) aaBBCc (3 dark) 1 2 AABBcc (4 dark) 1 AaBBcc (3 dark) 2 aaBBcc (2 dark) 1 AABbCC (5 dark) AaBbCC (4 dark) aaBbCC (3 dark) 2 4 2 AABbCc (4 dark) 4 AaBbCc (3 dark) 8 aaBbCc (2 dark) 4 AABbcc (3 dark) 2 AaBbcc (2 dark) 4 aaBbcc (1 dark) 2 phenotypes: 6 dark alleles 5 dark alleles 4 dark alleles 3 dark alleles 2 dark alleles 1 dark allele AAbbCC (4 1 dark) AAbbCc (3 dark) 2 AAbbcc (2 dark) 1 AabbCC (3 2 dark) AabbCc (2 dark) 4 Aabbcc (1 dark) 2 aabbCC (2 1 dark) aabbCc (1 dark) 2 aabbcc (0 dark) 1 0 dark alleles How many of each phenotype?