Population dispersion patterns in Ashe Juniper (Juniperus
advertisement

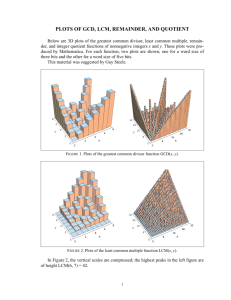
BIO 208L McMurry SPRING 2001 Population dispersion patterns in Ashe Juniper (Juniperus Ashei) In this lab, we will examine the dispersion pattern of a population of Juniperus Ashei, otherwise known as Ashe juniper. We will compare the results obtained by using plot and plotless methods. In central Texas, Ashe juniper is the dominant species of most upland communities. Its populations appear to be expanding, based on the relatively high percentages of the smallest size class (40-65%). This differs from the pattern shown in the other major deciduous tree species of the area, such as Texas oak (Quercus texana), Texas ash (Fraxinus texensis), Lacey oak (Quercus glaucoides), black cherry (Prunus serotina) and black walnut (Juglans nigra). These species show little evidence of recruitment into the smallest size classes (van Auken 1988). PLOT METHODS OF MEASURING DISPERSION The plot method involves setting up quadrats and then counting the number of juniper plant within each plot. The distribution of plots with varying numbers of trees should follow a Poisson distribution if the dispersion pattern is random (Equation 1). Equation 1. P(X) = e-X X! where = true mean number of individuals per plot for the entire pop, e = base of natural log (2.71828), and X = the number of individuals within a plot. X! means X factorial. For example, if X = 3, 3! = 3*2*1 = 6. Mu () is estimatedby X, the mean number of individuals per plot. When individuals are randomly dispersed within the population, the values of P(X) will be those predicted by the Poisson distribution. There is an equal and independent chance of a member of the population occurring at any point in the habitat. Testing if the dispersion pattern is significantly different from random There are several ways to test for nonrandom dispersion patterns using plots. We will examine three in more detail. Graphical comparison The observed dispersion pattern can be compared to the expected pattern if the population were randomly dispersed. From lab, we have a series of plots, each containing a number of juniper trees. Sum up the number of plots with varying numbers of junipers (column B) and divide by the total number of plots to determine the observed proportion of plots with 0-5 trees (column C). Table 1. Example of calculating the observed proportions of plots with X junipers. A B C 1 X # plots Observed 2 0 15 0.429 3 1 10 0.286 4 2 7 0.200 1 BIO 208L McMurry 5 3 3 0.086 6 4 0 0 7 5 0 0 8 Sum 35 1.00 9 Average 0.943 SPRING 2001 Next, calculate what we would expect if the trees followed a Poisson distribution, i. e., if they were randomly distributed. Use the Poisson function in Excel to do this. You may insert the function by using the paste function icon (fx) on the standard toolbar, by choosing "Insert….function" from the menu at the top of the screen, or by simply typing "=Poisson()" and filling in the required arguments within the parentheses. The Poisson function requires three arguments. The first argument is X, the number of individuals per plot. The second argument is X, the mean number of individuals per plot, calculated from our data. The third argument tells Excel if the cumulative probability should be calculated. We want to calculate the probabilities of each X separately, so for the third argument, type in "FALSE". Enter the formula for the first X (0) in cell D2: =POISSON(A2,$B$9,FALSE). "A2" refers to the cell with the X value, "$B$9" refers to the cell that contains the mean number of junipers per plot. Use the dollar signs to make sure that the rest of the formulas use the same value. Fill down across the values of X to get the Poisson values for those values (Table 2). 2 BIO 208L McMurry SPRING 2001 Table 2. Example of calculating the observed and expected proportions of plots with X junipers A B C D 1 X # plots Observed Expected 2 0 15 0.429 0.38951335 3 1 10 0.286 0.36725544 4 2 7 0.200 0.17313471 5 3 3 0.086 0.05441377 6 4 0 0 0.0128261 7 5 0 0 0.00241864 8 Sum 35 1.00 9 Average 0.943 The numbers in the expected and observed columns represent what we actually saw versus what we would have expected to see if the distribution of junipers is random. We can graph the two sets of numbers (Figure 1). In a random dispersion pattern, the observed proportion of plots with X junipers will be similar to that predicted by the Poisson distribution. In a uniform dispersion pattern, there will be more plots with few individuals (1-2) than the Poisson distribution predicts and fewer plots with more individuals. In a clumped dispersion pattern, there will be fewer plots with 1-2 individuals and more plots with more individuals than expected. The graphical examination method suffers from subjectivity as there is no test to determine if the observed pattern is significantly different from the expected pattern predicted by the Poisson distribution. To test for significant departures from randomness using plots, we must use either the variance-to-mean ratio, a goodness of fit test, or the Morisita Index. Variance to mean ratio One of the simplest methods of determining dispersion patterns using a plot method is to compare the variance to the mean. In a population having a Poisson distribution, the population mean, , is equal to the population variance, 2. Therefore, the variance to mean ratio should be 1.0. A ratio much less than 1.0 indicates a uniform dispersion, one much greater than 1.0 indicates a clumped dispersion. The population mean, , and the population variance, 2, are estimated by X and s2, respectively. The significance of the departure from a variance-mean ratio of 1.0 may be assessed by computing t as shown in Equation 2 and comparing it to the critical value of t for n-1 degrees of freedom in Table 1.B.1 in Brower et al. (1998), where n = the number of plots sampled. If the calculated value of t is less than the critical value, the dispersion pattern is random, even if the variance/mean ratio is different from 1. Equation 2. 3 BIO 208L McMurry SPRING 2001 2 s 1.0 X t 2 (n 1) Goodness of fit test The goodness of fit test used for determining if a dispersion pattern is random is the chi square (2 ) test. The observed frequencies of plots with X individuals is compared to those predicted by the Poisson distribution (Table 3). The observed frequencies (fx) are the number of plots with 0, 1, 2, etc., individuals. The expected frequencies (Fx) are determined by multiplying the expected proportions calculated in the graphical method by the total number of plots. Plots predicted to occur at a frequency less than one should be lumped together so that their cumulative frequencies are greater than one. Table 3. Goodness of fit test for random dispersion using the Poisson distribution. A B C D E X Observed Expected 1 fx (observed Fx (Expected frequency of plots) 2 3 4 5 6 7 0 1 2 3 4 5 15 10 7 3 0 0 0.429 0.286 0.200 0.086 0.000 0.000 0.3895 0.3673 0.1731 0.0544 0.01283 0.002419 frequency of plots) 13.6 12.9 6.1 1.9 0.45 0.08 In the example above, the expected frequencies (Fx)for 4 or more individuals are less than one, so they must be combined with the Fx for X = 3. Since some small proportion of plots would be expected to have 6 or more individuals, it is more accurate to sum the expected frequencies for the 0-2 categories and subtract the sum from the total number of plots to estimate the frequency of 3 or more individuals. Then the 2 statistic is calculated by plugging the expected and observed frequencies into the 2 formula in Equation 3. Equation 3. Using the above example, ( f F)2 2 F (15 13.6)2 (10 12.9)2 (7 6.1)2 (3 2.5) 2 1.02 13.6 12.9 6.1 2.5 The critical 2 value for alpha = 0.05 and 3 degrees of freedom is 7.815. Our calculated value (1.02) is much less than the critical value so the dispersion pattern is essentially random. 4 BIO 208L McMurry SPRING 2001 Morisita Index The variance-to-mean ratio and the goodness of fit test have a potential disadvantage in that they are affected by population size and plot size (Brower et al. 1998). An alternative plot method is Morisita’s index of dispersion (Id), which is not dependent on either parameter. Morisita’s index is shown in Equation 4. Equation 4. Id X n N N(N 1) 2 where n = the number of plots, N = the total number of individuals counted on all plots, X2 = the squares of the numbers of individuals per plot, summed across all plots. If Id = 1.0, the dispersion is random, if Id = 0, the population is perfectly uniform, and if Id > 1 the population is clumped. The degree of departure from randomness may be assessed statistically by computing Equation 5. Equation 5. 2 n X N 2 N where n = number of plots, X = number of individuals per plot, and N = number of individuals across all plots. The chi-square value may be compared to the critical value in Table 1.B.3 with n-1 degrees of freedom for the appropriate level (in this case, 0.05). 5 BIO 208L McMurry SPRING 2001 Plotless methods of measuring dispersion Plotless methods are usually less time-consuming than methods involving plots (Brower et al. 1998). Two plotless methods, the Holgate and Johnson-Zimmer methods, use point-to-plant distances, unlike nearest neighbor methods, which use plant-to-plant distances Point-to-plant distance methods may be superior to plant-to-plant distance methods due to the scale of nonrandom distributions. Non-random distributions may show patchiness on several levels of scale. For example, if plants are clumped, they may be arranged in clumps of clumps, rather than in simple clumps. An index based on plantto-plant distances will only show the smallest scale of nonrandomness present. One based on point to plant distances will be affected by most, if not all, the levels of nonrandomness in the population (Pielou 1965). The Holgate Method The Holgate method involves measuring point-to-plant distances for a set of randomly selected points. First, mark a set of randomly selected points (n). Then measure the distance from each point to the nearest plant (d) and the distance from the point to the second closest plant (d'). Then square both numbers and take the ratio for each point. Sum the ratios, divide by the number of points (n) and subtract 0.5 (Equation 6). Equation 6: d2 d' 2 0.5 n If the population is randomly dispersed, A will be 0; if it is clumped, A will be greater than 0; if it is uniformly dispersed, A will be less than 0. A If A is different from 0, you can test if the dispersion pattern is significantly different from random by computing the following test statistic (Equation 7) and comparing it to the critical value of t in Table 1B.1 of Brower et al. (1998) for n-1 degrees of freedom (n = the number of points sampled). Equation 7. A n 12 Payendah (1970) found that Holgate's ratio test produced reasonable results for computer simulated random and uniform distributions, but failed to detect nonrandomness for the original, semi-random and semi-uniform simulations. t Johnson and Zimmer method The Johnson and Zimmer method of calculating departures from random distribution is similar to the Holgate method. The main difference is that it only uses the distance from 6 BIO 208L McMurry SPRING 2001 the randomly selected point to the nearest plant, and, of course, the index itself is different. The index of aggregation is shown in Equation 8. Equation 8. 2 A (n 1) (di2 ) 2 2 ( di ) 2 As in the Holgate method, A = 0 indicates a random distribution, A>0 indicates a clumped distribution, and A<0 indicates a uniform distribution. Determining if departures from a random dispersion are statistically significant may be done by using Equation 9. Equation 9. t A 4(n 1) (n 2)(n 3) Compare the calculated value of t to the critical value of t for n-1 degrees of freedom (where n = number of points sampled) and =0.05. If it is less than the critical value, the population dispersion is not significantly different from random. If the calculated t value is greater than the critical value, then the population dispersion pattern is significantly different from random at the 0.05 level. Lab assignment We will select a stand of junipers at St. Edward’s Park. We will set up two transects perpendicular to each other. The exact length of each transect will be determined when we get to the site, but will be approximately 50-100 m in length. We will mark off the area at 10 m intervals to create a series of 10 x 10 m plots. Once the plots are in place, we will record the number of junipers within each plot and record that information. Working in teams, each team will randomly select a series of X and Y coordinates and mark those points . They will record the distance from each of their points to the closest and second-closest juniper. If the data are available, they will record the sex of the junipers. At the end of the lab, we will compile our data and it will be posted on the web for your use in completing the post-lab assignment. Post-Lab assignment The assignment from this lab is to complete the worksheet that accompanies this handout. It is worth 100 points and is due in two weeks. Literature Cited 7 BIO 208L McMurry SPRING 2001 Brower, J. E., J. H. Zar, and C. N. von Ende. 1998. Field and laboratory methods for general ecology. 4th ed. Boston: McGraw-Hill. 273 pp. Payendah. B. 1970. Comparison of method for assessing spatial distribution of trees. Forest Science 16:312-317. Pielou, E. C. 1959. The use of point-to-plant distances in the study of the pattern of plant populations. J. Ecol. 47:607-613. van Auken, O. W. 1988. Woody vegetation of the southeastern escarpment and plateau. In: Amos, B. B. and F. R. Gehlbach,, eds. Edwards Plateau vegetation: plant ecological studies in central Texas. Baylor Univ. Press, Waco, TX. p. 43-56. 8