structural bioinformatics
advertisement

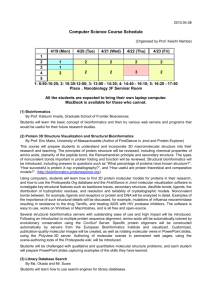
MSc in Bioinformatics
Module 2: Core Bioinformatics
Lesson 7. 3
Structural Bioinformatics
Molecular Modelling tools
Jean-Didier Maréchal
The Biotechnological Computational Chemistry Team
Department of Chemistry (UAB)
1
Course 2013-14
Module 2: Core Bioinformatics
MSc in Bioinformatics
General information
• JeanDi…
–
–
–
–
Email: jeandidier.marechal@gmail.com
Webpage: gent.uab.cat/jdidier
Room: C7/032 (chemistry building)
Research:
•
•
•
•
Enzyme design
Drug design (novel approaches for HIV, Al, Metabolism)
Peptide development
Software development
– Past: Academia, big pharma and spin off
– From computational chemistry to structural
bioinformatics
Structural Bioinformatics
Molecular Modelling tools
2
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
While the teacher bores me…
- Get your comp to linux
- Download the daily build of UCSF
Chimera
- Install it
Structural Bioinformatics
Molecular Modelling tools
3
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Introduction of Molecular Modeling
• Many atomic properties of the macromolecules can
not be experimentally assessed
• Molecular Modeling tools are key elements of
structural bioinformatics
• Molecular Modeling aims to provide with
reproductive and hopefully predictive simulations of
the molecular systems
• To do so, simulations are carried out with models that
explicitly represent the atoms in the molecular
system.
Structural Bioinformatics
Molecular Modelling tools
4
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Model I. Physics
• Molecular modeling studies lays
on the physical models used for
the atomic representation of the
systems
• The quality of the results is
directly proportional to the
exactness of the model
A physical model of a
reality
provides
Set of mathematic equations
defining the model
Applied on
a given system
• By default, the results provided by
modeling can not be exact
Solved through
computation
An estimated
behavior
Descriptive
Structural Bioinformatics
Molecular Modelling tools
5
Predictive
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Model II. Size does matter...
• Algorithms and computer or time
resources intrinsically limit the
size of the system that can be
treated
• Hence, modeling can also involves
the reduction of the number of
structural variables
– Study of only a part of the real
system
– Replacement of explicit solvent
molecules by a continuum
environment
– coarse grain approaches
– …
Structural Bioinformatics
Molecular Modelling tools
6
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
A model is ...a model
• The validity of a molecular modeling calculation
underlays in its approximations
–
–
–
–
Size
Environment
Physico-chemical conditions
…
• Results have to be discussed in the applicative
framework of the model:
– Do not over criticize the results
– Do not overstate the outcomes
Structural Bioinformatics
Molecular Modelling tools
7
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
The early XXIst Century Modeller
• Many models can be used to
simulate the atomic behavior of
molecules
• Each technique relies on its
approximation which its field of
applicability
• All of them are based on
estimating the energy of a given
spatial arrangement of atoms and
reach for the stables, metastables
and transition structures
Structural Bioinformatics
Molecular Modelling tools
8
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
The Potential Energy
Conformational Energy
•
•
Energy
( kcal /mol )
The potential energy, E, is a function of
the coordinates (positions), R, of all the
atoms in the system
60.0
42.0
24.0
5.0
-180
To a given geometry of the system
corresponds a unique value of potential
energy (if no electronic changes are
involved)
180
The entire map of the potential energy of
a system in function of its coordinates is
called the Potential Energy Surface (PES)
-90
-45
0
45
90
135
180
•
Because of the high dimensionality of the
entire PES, studies and analysis are
generally simplified to a reduced number
of variables
•
The question is how to calculate the
energy and how to explore the PES
9
Conformational Energy
4411.38
90
0
-90
-180
-180
Structural Bioinformatics
Molecular Modelling tools
-135
C(2)-C(4)-C(6)-C(11)(degrees)
N ( 7)- C ( 6)- C ( 5)- C ( 4)( degrees )
•
20.31
-90
0
90
C(5)-C(6)-N(7)-C(8)(degrees)
180
kcal/mol
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Some key questions for Molecular
Modeling
• Characterization of the most stables
conformations of a system
• Atomic description of dynamical properties
of the protein
• Determination in silico of the structural
features of protein (i.e. Homology
Modeling)
• Decode nature of interactions between
biomolecules
• Determination of the catalytic processes
Structural Bioinformatics
Molecular Modelling tools
10
Some examples
in this lesson
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Calculation of the energy
Structural Bioinformatics
Molecular Modelling tools
11
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Molecular events
Change in chemical state
Fiting/binding
Pre-organization
product
Affine chemical compoud
Good sampling
Accurate electronic
Structural Bioinformatics
Molecular Modelling tools
12
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
The three main molecular modeling families
13
Structural Bioinformatics
Molecular Modelling tools
13
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
A - Quantum techniques
•
Generally solve Schrödinger equation…
–
–
•
time independent
in the Born-Oppenheimer approximation (electronic PES)
Implies that the structure with the lowest energy is the most occupied over time
( x, y, z) ( x, y, z)
Hamiltonian
Energy
Wave function
With the exact hamiltonian:
N
ˆ
A1
Z Z
2M
R
1
2
A
A
N
N
A
i 1
j i
n
n
n
N
1
2
1
i
Z A
i 1 2
i 1 j i r ij
i 1 A1 r iA
n
B
AB
1
Structural Bioinformatics
Molecular Modelling tools
Tn
Vnn
14
Te
Vee
14
V
en
Jean-Didier
Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
QM techniques
•
Different levels:
– ab initio
– huckel, extended huckel, semi empiric..
– Functional Density Theory
•
Techniques used when aiming to high quality results
•
Necessary for processes with changes in electronic nature of the system:
– Catalysis
– Changes in covalent bonds
– Changes in coordination bonds
•
QM method still have a substantially high ratio Time/natom
Structural Bioinformatics
Molecular Modelling tools
15
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
B - Molecular Mechanics techniques
• Nuclei and electrons behaviors
incorporated in a potential
l
• Parametrization of the different
kind of atomic forces
VEnllaç kb (l l0 ) 2 VPlegament kb ( 0 ) 2
d-
• Will be necessary to treat
conformational changes large
molecular system
VElectrostatic
i
• Can not treat changes in chemical
natures
Structural Bioinformatics
Molecular Modelling tools
d+
16
j i
qi q j
rij Vtorsió A[1 cos(n )]
VVDW 4 ij [( ij / rij )12 ( ij / rij )6 ]
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
QM .vs. MM
• At same number of atoms, the conformational
explorations are a lot faster for MM than QM
approaches
• This velocity of calculation allows to treat easily
the system time dependently with MM
• Some techniques allow to explore large
conformational motions
• MM can not treat changes in the chemical state
of the system
Structural Bioinformatics
Molecular Modelling tools
17
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
C – QM/MM methods
•
Enzymes performs catalysis at their active
site
•
The proteic environment has some impact
on the active centre:
– Steric
– Electrostatic
– …
•
When modeling the entire system QM and
MM approximations are required
•
Hybrid QM/MM
– Part of the protein is treated with MM
techniques
– Where the key region is located, QM is used
(example catalytic center)
Structural Bioinformatics
Molecular Modelling tools
18
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Folding
Sampling
Recognition
Catalysis
Fine
Electronics
=
Quantum
Based
Structural Bioinformatics
Molecular Modelling tools
Docking
Homology
Modeling
Normal Mode
Analysis
Wide
space
=
Approx.
Energy
Molecular
Dynamics
QM and QM/MM
Motions
Transition
metal
19
Simplicity
of calculation
of E
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Exploration of the PES
Structural Bioinformatics
Molecular Modelling tools
20
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
A multidimensional problem
The number of local minima typically increases exponentially with the number
of variables (degrees of freedom).
Possible Conformations (3n) for linear
alkanes CH3(CH2)n+1CH3
n=1
n=2
n=5
n = 10
n = 15
• Combinatorial Explosion Problem n = 100
Structural Bioinformatics
Molecular Modelling tools
21
3
9
243
59,049
14,348,907
?
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Minimization
E
?
Transition
state
?
?
Local
Minimum
Global
Minimum
r
22
Structural Bioinformatics
Molecular Modelling tools
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Minimization: General Scheme
1.
2.
3.
4.
start at an initial point and
calculate E
determine according to a fixed
rule a direction of movement
Coordinates {x}0
Energy
move in that direction to a
(hopefully)
lowest
energy
structure.
At the new point, a new
direction is determined and the
same process is repeated.
The primary difference between
algorithms is the rule by which
successive directions of movement are
selected.
Structural Bioinformatics
Molecular Modelling tools
NO
Gradient
Hessian
Search Algorithm
Converged?
YES
Optimized
New coordinates {x}1
23
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Optimizers main families
• Don’t have any derivatives (hard to know which way to
move to reduce function value)
– Simplex method
– Sequential Univariant Method
Not efficient
• Do have derivatives (use them to move toward minimum )
– Line optimization
• Golden Mean Method
• Parabolic Optimization
– First derivate methods
• Steepest Descent
• Conjugate Gradient
– Second derivative methods
• Newton-Raphson
Structural Bioinformatics
Molecular Modelling tools
24
ef
1
ef
f ( x) f2
e3
f ( x d e1 ) f ( x )
f ( x d e2 ) f ( x )
d
f ( x d e3 ) f ( x )
d
d
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Steepest Descent
Data:
x0 R
n
Step 0: set i=0
Step 1: if
f ( xi ) 0
stop
hi f ( xi )
i arg min f ( xi hi )
else, compute search direction
Step 2: compute the step-size
Step 3: set
Structural Bioinformatics
Molecular Modelling tools
xi 1 xi i hi
25
0
go to step 1
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Conjugate Gradient
• The basic idea: decompose the n-dimensional
quadratic problem into n problems of 1dimension
• This is done by exploring the function in
“conjugate directions”
• CG will find minimum of an N-dimensional
quadratic function in at most N steps! Nonquadratic functions take longer – but all functions
become quadratic near their minimum so CG is
efficient
Structural Bioinformatics
Molecular Modelling tools
26
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Practical aspects
• SD are generally applied at the beginning of the optimization and the CG at the
end.
• Other methodologies are even more eficient when one want to reach accuracy
in the determination of the minimium (Newton-Raphson)
• In many cases, it could be interesting to start with different structures.
• And to verify (frequency) that we are indeed with a minimum.
• For macromolecules:
• Minimization is used to relax the structure but not to catch the exact absolute minimum
• The minimization generally ends in a local minimum
Structural Bioinformatics
Molecular Modelling tools
27
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Exercise 1.
Lets get minimized
Minimization cyclosporin A
Structural Bioinformatics
Molecular Modelling tools
28
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
From wells to wells
T
R
Structural Bioinformatics
Molecular Modelling tools
29
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
More than a structure
• Minimization only provides one structure: the
closest minimum of a given starting point
• As the degrees of freedom of system increase,
the number of minima increase
• Exploring the PES is not as trivial
• Numerous methodologies aim at exploring the
conformational space:
– To locate the best minimum
– To extract statistical data with thermodynamical
means
Structural Bioinformatics
Molecular Modelling tools
30
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Some common conformational
exploration schemes
• Monte Carlo: Allows random changes of the
structure and evaluate their energetical cost. Low
energy structures are kept (can form ensembles
that are statistically relevant)
• Genetic Algorithms: Structural displacements mix
randoms and evolutionary guided changes. Only
low energy structures are kept based on survival
criteria
• Simulated Annealing: Overheat the system to
allow barrier jump then cool down to encounter
lowest energy structures
Structural Bioinformatics
Molecular Modelling tools
31
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Molecular Dynamics
• Calculate the motion of the
atoms using Newtonian
dynamics
• determine the net force
and acceleration
experienced by each atom.
• Several algorithms are
used to calculate
displacements of the atom
over time (verlet,
leapfrog…)
• Like MC allow statistical
analysis
Structural Bioinformatics
Molecular Modelling tools
32
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Time steps
• Knowledge of the atomic forces and masses can be used to
solve the position of each atom along a series of extremely
small time steps (on the order of femtoseconds = 10-15
seconds).
• The resulting series of snapshots of structural changes over
time is called a trajectory.
Structural Bioinformatics
Molecular Modelling tools
33
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Time scale
Biological molecules exhibit a wide range of time scales
over which specific processes occur;
for example
Local Motions (0.01 to 5 Å, 10-15 to 10-1 s)
Atomic fluctuations
Sidechain Motions
Loop Motions
Rigid Body Motions (1 to 10Å, 10-9 to 1s)
Helix Motions
Domain Motions (hinge bending)
Subunit motions
Large-Scale Motions (> 5Å, 10-7 to 104 s)
Helix coil transitions
Dissociation/Association
Folding and Unfolding
Structural Bioinformatics
Molecular Modelling tools
34
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
The meaning of trajectories
Vibrations in proteins
vary widely in energy
Low frequencies
vibration correspond
to collective motion
of the proteins
High frequencies
vibration to localized
motions
Structural Bioinformatics
Molecular Modelling tools
35
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Relationship accuracy / time scale
• Computing numerous
structures (energy, gradient,
forces, etc.) is increasingly
ressource demanding in
function of the quality of the
energetic model
• Force field approaches are
simplified enough so that
calculations can be performed
on a very wild conformational
and chemical space
• Simulations can be performed
nowadays on solvated systems
and for long runs
Structural Bioinformatics
Molecular Modelling tools
36
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Exercise 2
Molecular Dynamics of Cyclosporin A
Structural Bioinformatics
Molecular Modelling tools
37
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Homology Modeling
Structural Bioinformatics
Molecular Modelling tools
38
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
The problem
•
Most biochemical projects
(drug design, enzyme design,
etc.) require the physical
three dimensional
structure of the
physiological target
•
Experimental resolution
(NMR or X-ray) is not always
accessible.
•
Computational tools have
been set up to produce
models of proteins to further
study
– Ab initio
– Comparative/homology
modeling
Structural Bioinformatics
Molecular Modelling tools
39
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
The grounds of comparative modeling
• From the mid 80s, studies showed that:
– Proteins with SeqID upper 80% mainly have
differences in fold in the range of experimental error
– Up to 30-20%, protein share a strong structural
similarity
– Below this threshold protein might be or not
structurally related.
• With a good engouh SeqID and alignment
modeling could find out its way to produce
accurate models.
Structural Bioinformatics
Molecular Modelling tools
40
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
The general methodology
Required material:
The structure of a “close”
parent
The sequence of the target
protein
A sequence alignment
program (e.g. ClustalW, TCoffee)
A homology modeling
program (e.g. modeller)
Structural Bioinformatics
Molecular Modelling tools
41
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
A framework like modeller
Step 1 and 2: research of templates
1. template recognition
and sequence alignements
Step 3 – Generation of
2. alignment
main chain model
3. alignment correction
4. backbone generation
5. generation of canonical loops
Step 4 and 5 – Optimization of side chains and flexible parts
(data based)
6. side chain generation plus
optimisation
7. ab initio loop building (energy
based)
8. overall model optimisation
(energy minimisation)
step 6 – Full relaxation
9. model verification with
optional repeat of previous steps.
Structural Bioinformatics
Molecular Modelling tools
42
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
The success and the limitations
• When SeqID high, the method is generally highly
efficient.
• In dark regions or difficult structural assignment,
homology modeling could be helped by
secondary structure prediction programs
• Moreover, multiple alignment can be particularly
useful
• HM methods are generally updated tools that
improve the evaluation of the quality of the
model and better explore the conformational
space of flexible regions (i.e. loops)
Structural Bioinformatics
Molecular Modelling tools
43
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Post Modeling
• The accuracy of the model has to be checked.
• Generally the same than those of experimental
structure
• Procheck(http://biotech.embl-ebi.ac.uk:8400/)
• Check for protein stereochemistry
– MolProbity (http://molprobity.biochem.duke.edu/)
• Ramachandran plot, bond length etc
– Verify3D (http://www.doembi.ucla.edu/Services/Verify_3D/)
• Check sequence vs structure
Structural Bioinformatics
Molecular Modelling tools
44
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Exercise 3
Not quite!
Structural Bioinformatics
Molecular Modelling tools
45
Jean-Didier Maréchal
Module 2: Core Bioinformatics
MSc in Bioinformatics
Folding
Sampling
Recognition
Catalysis
Fine
Electronics
=
Quantum
Based
Structural Bioinformatics
Molecular Modelling tools
Docking
Homology
Modeling
Normal Mode
Analysis
Wide
space
=
Approx.
Energy
Molecular
Dynamics
QM and QM/MM
Motions
Transition
metal
46
Simplicity
of calculation
of E
Jean-Didier Maréchal