lac - Kustu Lab
advertisement

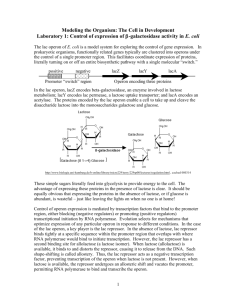
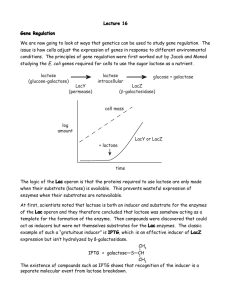
Periods 9, 10 and 11 (Apr. 2, 9, and 16) A) Induction of the lactose (lac) operon of E. coli and determination of the differential rate of synthesis of β-galactosidase, the product of the lacZ gene. Background: As you know, not all of the genes of E. coli are expressed at high levels under all growth conditions. The purpose of this experiment is to follow the kinetics of induction of the lactose operon, in particular the lacZ gene, which codes for the enzyme β-galactosidase. We will determine the differential rate of synthesis of β-galactosidase, that is the proportion of the total protein being synthesized that is constituted by this enzyme. To induce the lac operon, we will draw on an observation of Jacob and Monod, who also discovered operons. Namely, the compound isopropyl-β-D-thiogalactoside (IPTG—see p. 8-8 of your manual) is bound by the lactose repressor, the product of the lacI gene, and decreases the affinity of the repressor for the lactose operator. However, IPTG is not cleaved by β-galactosidase and so its concentration in the medium remains constant. Compounds like IPTG are called gratuitous inducers. (IF YOU DID NOT USE A GRATUITOUS INDUCER, HOW WOULD YOU INDUCE EXPRESSION OF THE lac OPERON?) We will grow E. coli on enriched medium (LB), simply because it grows rapidly and we can complete growth and sample collection during one lab period. After we have established that the cells are in the exponential growth phase, we will add IPTG to induce expression of the lac operon to high levels. We will assay β-galactosidase during the next lab period. Materials: 1. A fully-grown culture of E. coli K12 on Luria Broth medium (5 ml/pair) 2. Luria Broth (100 ml in a sterile 500 ml or 1-liter flask) (1 per 2 pairs.) 3. Isopropylthiogalactoside (IPTG) (10 mM) (1 ml/pair) 4. Shaking water baths at 37°C with the appropriate holders for 500 ml or 1-liter flasks 5. Spectrophotometers warmed up 6. Cuvettes for spectrophotometers 7. Means of washing cuvettes between readings 8. Sterile pipettes 9. Buckets of ice 10. Graph paper (semi-log, provided in lab manual) 11. Vortex mixers 12. Sterile Eppendorf tubes 13. Test tubes and test tube racks for taking samples PMB C112L Spring 2007 Sydney Kustu 9-1 Methods: 1. Inoculate your LB culture at a 1/100 dilution. 2. Follow the O.D.650 of the culture in one of the spectrophotometers and allow approximately 2 doublings so that the cells are in mid-exponential growth. Plot your O.D. readings as a function of time so you will know this. It will be convenient to sample at intervals of one doubling time. 3. Just before adding IPTG, remove a 5-ml sample to a chilled test tube so that you can measure β-galactosidase activity next period. 4. Add IPTG to your remaining culture to a final concentration of 100 µM. (Take account of the amount of culture you have removed for O.D. readings.) (HOW MUCH OF THE 10 mM STOCK SOLUTION OF IPTG WILL YOU USE?) 5. Continue to follow the growth of the culture and, as you do, also remove 2.5-ml samples to ice for measurements of β-galactosidase activity next period. It will be convenient to sample at intervals of 20 minutes. 6. When your growth curve and sampling are completed, save the remainder of your culture for practice assays next period. Divide the pre-IPTG sample into several Eppendorf tubes so you save all of it. Divide the post-IPTG samples into two portions of ≥1 ml in Eppendorf tubes. LABEL ALL TUBES WITH GROUP (G) NUMBERS. 7. Save the starter culture to confirm that a fully-grown LB culture has not induced expression of the lac operon to high levels. Questions: 1. What is an operon? 2. Under what natural conditions is the lactose (lac) operon of E. coli highly expressed? Of what advantage is it to E. coli to regulate expression of the lac operon? 3. Is the lactose (lac) operon of E. coli regulated negatively or positively? How do you know? 4. Of what advantage to us are gratuitous inducers? PMB C112L Spring 2007 Sydney Kustu 9-2 B) Growth of phage P1. Background: See material for period 7 beginning on p. 7-1. Materials: 1. The same fully grown culture of E. coli K12 used above. The 5 ml/pair above will be plenty for this, too. 2. P1vir stock phage (0.2 ml/pair). (See attached for preparation). This should go in an ice bucket. 3. P1 broth (LB containing 0.2% glucose and 5 mM CaCl2 ) (10 ml/pair) 4. Test tube rack in one of the 37°C shakers 5. Bottle of chloroform in the hood 6. Pasteur pipettes 7. Test tubes and test tube racks for taking samples Methods: 1. Inoculate E. coli K12 into 5 ml of P1 broth at a 1/100 dilution. Inoculate a second tube to use as a control and label it bacterial control. 2. At an O.D.650 of approximately 0.3 to 0.4 (3 doublings), add 0.1 ml of P1vir stock phage to your experimental culture (but not your control culture). 3. Incubate for 2 to 3 hours with shaking at 37°C until you see lysis in the experimental culture (it will clear). The clearing of the experimental culture should be apparent by comparison to the control culture. 4. Add 4 or 5 drops of chloroform to the experimental culture and vortex for 30 sec. 5. Chill for use in a future transduction experiment. PMB C112L Spring 2007 Sydney Kustu 9-3 Preparation of P1vir phage stock for long term storage: 1. Inoculate 1 ml of a fully grown E. coli K12 culture into 100 ml of P1 broth. 2. At an O.D.650 of approximately 0.3 to 0.4 (3 doublings), add 2 ml of P1vir stock phage (i.e. phage that has not been concentrated. Dilute a concentrated storage stock appropriately more.) 3. Incubate 2 to 3 hours with shaking at 37°C until you see lysis. 4. To precipitate phage, add 2.5 g NaCl and 8 g PEG 6000* (see below). 5. After the PEG dissolves, centrifuge in a bottle for 15 min at 6000 rpm in the Sorvall. 6. Pour off the supernatant (save it on ice until you are sure the phage has pelleted). 7. Centrifuge the pellet briefly a second time so that you can remove more supernatant. Use a Pasteur pipette. 8. Suspend the pellet in 10 ml P1 storage buffer (10 mM Tris Cl, pH 7.5; 10 mM MgCl2; 1% ammonium acetate). 9. If the solution is viscous, add a crystal of DNase and incubate about 1 min at 37°C until the viscosity drops. 10. Add 10 ml of chloroform and roll gently by hand to make an emulsion. 11. Centrifuge for 5 min at 8000 rpm in the Sorvall to separate the phases. 12. Take the top phase and label it P1 storage stock. (13. If you want to remove even more medium components, you can dialyze the phage against P1 storage buffer, but this shouldn't be necessary for us.) * PEG 6000 from Sigma or USB. Sigma 8125B Biochemika Ultra for molecular biology. PMB C112L Spring 2007 Sydney Kustu 9-4