Conversion Tables and Formulas
advertisement
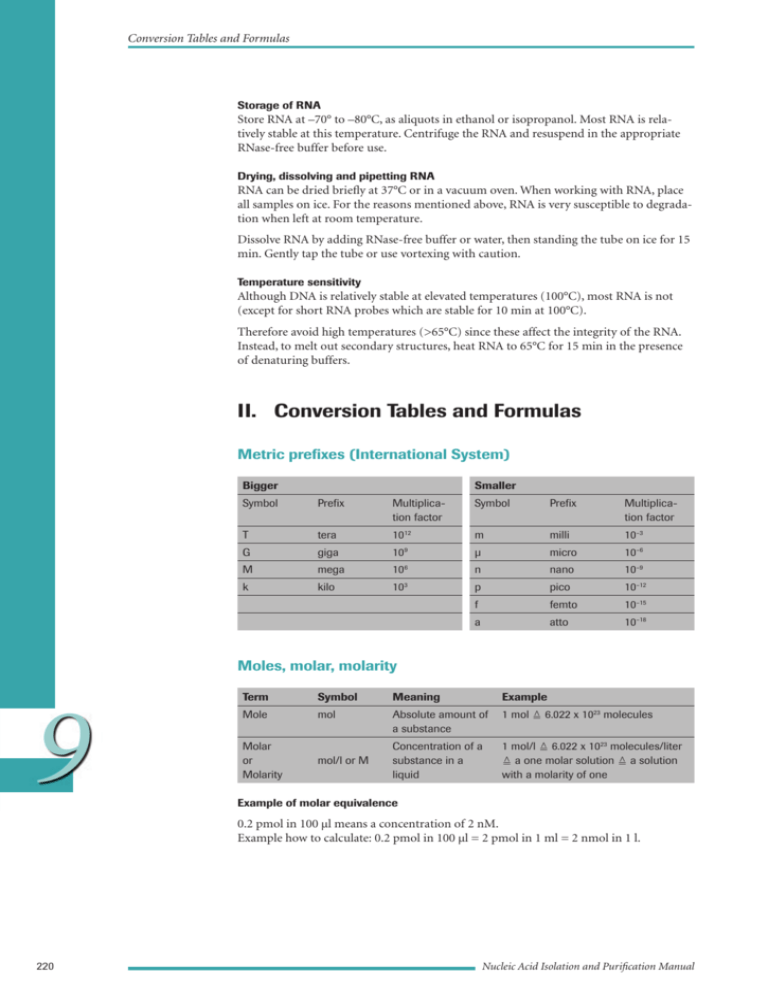
Conversion Tables and Formulas Storage of RNA Store RNA at –70° to –80°C, as aliquots in ethanol or isopropanol. Most RNA is relatively stable at this temperature. Centrifuge the RNA and resuspend in the appropriate RNase-free buffer before use. Drying, dissolving and pipetting RNA RNA can be dried briefly at 37°C or in a vacuum oven. When working with RNA, place all samples on ice. For the reasons mentioned above, RNA is very susceptible to degradation when left at room temperature. Dissolve RNA by adding RNase-free buffer or water, then standing the tube on ice for 15 min. Gently tap the tube or use vortexing with caution. Temperature sensitivity Although DNA is relatively stable at elevated temperatures (100°C), most RNA is not (except for short RNA probes which are stable for 10 min at 100°C). Therefore avoid high temperatures (>65°C) since these affect the integrity of the RNA. Instead, to melt out secondary structures, heat RNA to 65°C for 15 min in the presence of denaturing buffers. II. Conversion Tables and Formulas Metric prefixes (International System) Bigger Smaller Symbol Prefix Multiplication factor Symbol Prefix Multiplication factor T tera 1012 m milli 10–3 G giga 10 9 µ micro 10–6 M mega 106 n nano 10–9 k kilo 103 p pico 10–12 f femto 10–15 a atto 10–18 Moles, molar, molarity 9 Term Symbol Meaning Example Mole mol Absolute amount of a substance 1 mol 6.022 x 1023 molecules Molar or Molarity mol/l or M Concentration of a substance in a liquid 1 mol/l 6.022 x 1023 molecules/liter a one molar solution a solution with a molarity of one Example of molar equivalence 0.2 pmol in 100 µl means a concentration of 2 nM. Example how to calculate: 0.2 pmol in 100 µl = 2 pmol in 1 ml = 2 nmol in 1 l. 220 Nucleic Acid Isolation and Purification Manual Conversion Tables and Formulas Physical conversions and formulas Centrifugal force conversion (Dyson, 1991) RCF = (1.11 x 10–5) (rpm)2 r RCF = relative centrifugal force (x g; where g = 980 cm x sec–2) rpm = rounds per minute r = radius of rotor in mm Nucleic acid data Conversions Spectral constants for nucleotides (Adapted from Sambrook, et al., 1989) Compound Molecular weight (MW) max (pH 7.0) Absorbance at max (1 M solution) ATP 507.2 259 15,400 dATP 491.2 259 15,400 CTP 483.2 271 9,000 dCTP 467.2 272 9,100 GTP 523.2 253 13,700 dGTP 507.2 253 13,700 UTP 484.2 260 10,000 dTTP 482.2 267 9,600 Molar concentration of nucleic acid = (observed absorbance at max) ÷ absorbance at max for 1 M solution. Spectrophotometric equivalents 1 A260 unit Nucleic acid Amount Molarity (in nucleotides) double-stranded DNA 50 µg/ml 0.15 mM single-stranded DNA 33 µg/ml 0.10 mM single-stranded RNA 40 µg/ml 0.11 mM oligonucleotide* 20 – 30 µg/ml 0.06 – 0.09 mM * For exact determination of the molecular weight, see table “Conversions between weight and molarity of various DNAs” page 223. Calculations 9 Determining purity of nucleic acid preparations For pure DNA: A260/A280 1.8 For pure RNA: A260/A280 2.0 An A260/A280 ratio of <1.8 (DNA) or <2.0 (RNA) means the nucleic acid preparation contains contaminations (e.g., protein), or phenol. Appendix 221 Conversion Tables and Formulas Calculating molecular weight of nucleic acids (Adapted from Ausubel et al., 1988) For molecular weight of Use this calculation DNA base pair (sodium salt) 1 base pair = 665 daltons double-stranded DNA molecule (number of base pairs) x (665 daltons/base pair) single-stranded DNA molecule (number of bases) x (325 daltons/base) single-stranded RNA molecule (number of bases) x (340 daltons/base) oligonucleotide For dephosphorylated oligonucleotides: [(number of A x 312.2) + (number of G x 328.2) + (number of C x 288.2) + (number of T x 303.2)] – 61 For phosphorylated oligonucleotides: [(number of A x 312.2) + (number of G x 328.2) + (number of C x 288.2) + (number of T x 303.2)] + 17 Conversions between picomoles and micrograms of DNA For double-stranded DNA (dsDNA): To convert Calculate* pmol to µg pmol N µg to pmol µg 1µg 660pg = µg 1pmol 106pg pmol 106pg = pmol 1µg 660pg * N = number of base pairs in DNA; 660, average molecular weight of a base pair. For single-stranded DNA (ssDNA): To convert Calculate* pmol to µg pmol N µg to pmol µg 1µg 330pg = µg pmol 106pg pmol 106pg 1 = pmol N 1µg 330pg * N = number of nucleotides in DNA; 330, average molecular weight of a nucleotide 9 Calculating moles of ends Moles of ends for double-stranded DNA molecule: 2 x (grams of DNA) / (MW in daltons) Picomoles of ends per microgram of double-stranded DNA: (2 x 106) / (660 x number of bases) Moles of ends generated by restriction endonuclease cleavage, circular DNA molecule: 2 x (moles of DNA) x (number of sites) Moles of ends generated by restriction endonuclease cleavage, linear DNA molecule: [2 x (moles of DNA) x (number of sites)] + [2 x (moles of DNA)] 222 Nucleic Acid Isolation and Purification Manual Conversion Tables and Formulas Conversions between weight and molarity of various DNAs Type Size pmol/µg Molecules/µg oligonucleotide 20 nucleotides 152 µg/pmol 9.1 x 10 13 0.0066 11 0.66 DNA 1000 bp 1.52 9.1 x 10 pUC18/19 DNA 2686 bp 0.57 3.4 x 1011 1.77 pBR322 DNA 4361 bp 0.35 2.1 x 10 11 2.88 M13mp18/19 DNA 7250 bp 0.21 1.3 x 10 11 4.78 48,502 bp 0.03 1.8 x 1010 32.01 DNA Protein data Conversion between DNA and protein 1 kb of DNA = 333 amino acids of coding capacity = a protein of 3.7 x 104 daltons Conversion between protein molecular weight and absolute weight 100 pmol MW of protein [kdaltons] Amount in µg 100 10 50 5 10 1 9 Appendix 223




