Macromolecular Interactions of Core and Linker Histone 'tail'
advertisement

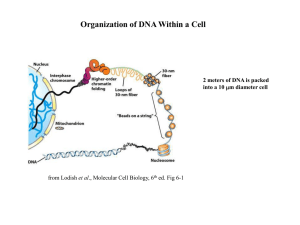
Department of Chemistry and Biochemistry Seminar Announcement October 22, 2015 Room 114 Smith Hall, 12:30 p.m.* Dr. Jeffrey Hayes University of Rochester Medical Center Department of Biochemistry and Biophysics Macromolecular Interactions of Core and Linker Histone 'tail' Domains that Control Chromatin Structure and Function The chromosomes of humans and other eukaryotes provide the seemingly contrasting functions of compacting the genomic DNA several thousand times its length while also allowing for efficient processes such as replicating the genome and expressing genes encoded within. To accomplish this, the DNA is assembled into a complicated, multifaceted complex known as chromatin. The initial level of packaging DNA into chromatin involves wrapping short ~200 bp segments around protein spools comprised of the core histone proteins into structures known as nucleosomes. Immensely long, genome-sized strings of nucleosomes are assembled into a hierarchy of higher-order chromatin structures, to form chromosomes. Formation of such higher-order involves essential inter-nucleosome interactions provided by the ‘tail’ domains of the core histone proteins, which protrude out from the main body of nucleosomes. Posttranslational modification within these domains is a key component in the regulation of gene expression. While nucleosome structure is fairly well understood, higherorder chromatin structures and inter-nucleosome interactions remain poorly defined. Moreover, how posttranslational modifications within the tail domains result in modulation of chromatin higher order structures to regulate gene expression is not well understood. Using site-directed crosslinking, we have defined intra- and inter-nucleosome interactions for core histone tail domains and how an additional chromatin component, linker histone H1, affects these contacts. In addition, using biochemical and fluorescence techniques, we have defined the COOH-terminal domain (CTD) of linker histones as an intrinsically disordered domain and shown that the CTD adopts defined structure(s) upon H1 binding to nucleosomes. Kan, P.-Y., Lu, X, Hansen, J.C. and Hayes, J.J. (2007). The H3 tail domain participates in multiple interactions during folding and selfassociation of nucleosome arrays. Mol. Cell. Biol. 27, 2084-2091. Wang, X, and Hayes, J.J. (2008). Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating stability of higher-order chromatin structure. Mol. Cell. Biol. 21, 227–236. Kan, P.-Y., Caterino, T., and Hayes, J.J. (2009) The H4 Tail Domain Participates In Intra- And Inter-Nucleosome Interactions With Protein And DNA During Folding And Oligomerization Of Nucleosome Arrays. Mol. Cell Biol. 29, 538-546. Caterino, T.L., Fang, H., and Hayes, J.J. (2011). Nucleosome Linker DNA Contacts and Induces Specific Folding of the Intrinsically Disordered H1 Carboxyl Terminal Domain. Mol. Cell Biol. 31:2341-2348. PMID: 21464206. Fang, H., Clark, D.J., Hayes, J.J. (2012). DNA and nucleosomes direct distinct folding of a linker histone H1 C-terminal domain. Nuc. Acids Res. 40:1475-1484 PMID: 22021384. PMC3287190 *Pre-seminar social time – 12:15 p.m. A pizza lunch with the speaker will follow after the presentation in the Smith Hall student lounge.