Factor Analysis Tutorial: Subject Marks & Athletics Data
Factor Analysis - 2
nd
TUTORIAL
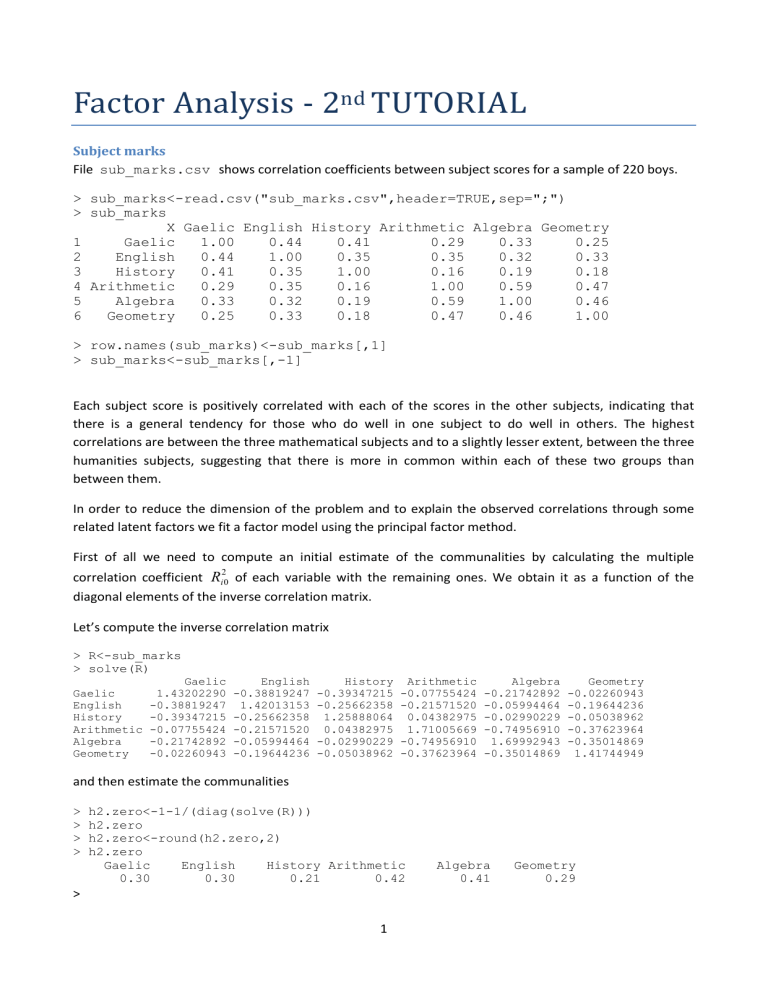
Subject marks
File sub_marks.csv shows correlation coefficients between subject scores for a sample of 220 boys.
> sub_marks<-read.csv("sub_marks.csv",header=TRUE,sep=";")
> sub_marks
X Gaelic English History Arithmetic Algebra Geometry
1 Gaelic 1.00 0.44 0.41 0.29 0.33 0.25
2 English 0.44 1.00 0.35 0.35 0.32 0.33
3 History 0.41 0.35 1.00 0.16 0.19 0.18
4 Arithmetic 0.29 0.35 0.16 1.00 0.59 0.47
5 Algebra 0.33 0.32 0.19 0.59 1.00 0.46
6 Geometry 0.25 0.33 0.18 0.47 0.46 1.00
> row.names(sub_marks)<-sub_marks[,1]
> sub_marks<-sub_marks[,-1]
Each subject score is positively correlated with each of the scores in the other subjects, indicating that there is a general tendency for those who do well in one subject to do well in others. The highest correlations are between the three mathematical subjects and to a slightly lesser extent, between the three humanities subjects, suggesting that there is more in common within each of these two groups than between them.
In order to reduce the dimension of the problem and to explain the observed correlations through some related latent factors we fit a factor model using the principal factor method.
First of all we need to compute an initial estimate of the communalities by calculating the multiple correlation coefficient
R 2 i 0
of each variable with the remaining ones. We obtain it as a function of the diagonal elements of the inverse correlation matrix.
> R<-sub_marks
> solve(R)
Gaelic English History Arithmetic Algebra Geometry
Gaelic 1.43202290 -0.38819247 -0.39347215 -0.07755424 -0.21742892 -0.02260943
English -0.38819247 1.42013153 -0.25662358 -0.21571520 -0.05994464 -0.19644236
History -0.39347215 -0.25662358 1.25888064 0.04382975 -0.02990229 -0.05038962
Arithmetic -0.07755424 -0.21571520 0.04382975 1.71005669 -0.74956910 -0.37623964
Algebra -0.21742892 -0.05994464 -0.02990229 -0.74956910 1.69992943 -0.35014869
Geometry -0.02260943 -0.19644236 -0.05038962 -0.37623964 -0.35014869 1.41744949 and then estimate the communalities
> h2.zero<-1-1/(diag(solve(R)))
> h2.zero
> h2.zero<-round(h2.zero,2)
> h2.zero
Gaelic English History Arithmetic Algebra Geometry
0.30 0.30 0.21 0.42 0.41 0.29
>
1
Now we can compute the reduced correlation matrix by substituting the estimated communalities to the diagonal elements (the 1's) of the original correlation matrix.
> R.psi<-R
> i<-1
> for (i in 1:nrow(R.psi)){
+ R.psi[i,i]<-h2.zero[i]
+ }
> R.psi
Gaelic English History Arithmetic Algebra Geometry
Gaelic 0.30 0.44 0.41 0.29 0.33 0.25
English 0.44 0.30 0.35 0.35 0.32 0.33
History 0.41 0.35 0.21 0.16 0.19 0.18
Arithmetic 0.29 0.35 0.16 0.42 0.59 0.47
Algebra 0.33 0.32 0.19 0.59 0.41 0.46
Geometry 0.25 0.33 0.18 0.47 0.46 0.29
>
R.psi is still squared and symmetric, but it is not positive definite. Its decomposition through the spectral theorem shows that only two eigenvalues are positive
> eigen(R.psi)
$values
[1] 2.06689350 0.43185860 -0.07389683 -0.11987084 -0.17312229 -
0.20186214
$vectors
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] -0.3894948 -0.4631310 -0.3323422 -0.08042293 0.49656626 0.5199098
[2,] -0.4068092 -0.3304872 0.4836944 -0.57629533 0.02029114 -0.3984924
[3,] -0.2898160 -0.5460418 -0.1179882 0.51251652 -0.56060428 -0.1642360
[4,] -0.4659483 0.4148356 -0.1362116 -0.32995348 -0.55776080 0.4150706
[5,] -0.4674485 0.3627504 -0.4779927 0.12767051 0.27396261 -0.5745187
[6,] -0.4039689 0.2728550 0.6282010 0.52304263 0.22930424 0.2038842 eigen.va<-eigen(R.psi)$values eigen.ve<-eigen(R.psi)$vectors
This means that two factors might be enough in order to explain the observed correlations.
The estimate of the factor loading matrix will then be obtained as , where has in columns the first two eigenvectors and
L
1
has on the diagonal the first two eigenvalues.
> l.diag
[,1] [,2]
[2,] 0.000000 0.4318586
> gamma<-eigen.ve[,1:2]
> gamma
[,1] [,2]
[1,] -0.3894948 -0.4631310
[2,] -0.4068092 -0.3304872
[3,] -0.2898160 -0.5460418
[4,] -0.4659483 0.4148356
[5,] -0.4674485 0.3627504
[6,] -0.4039689 0.2728550
2
We can now compute the estimated factor loadings:
> lambda<-gamma%*%sqrt(l.diag)
> round(lambda,2)
[,1] [,2]
[1,] -0.56 -0.30
[2,] -0.58 -0.22
[3,] -0.42 -0.36
[4,] -0.67 0.27
[5,] -0.67 0.24
[6,] -0.58 0.18
The first factor seems to measure overall ability in the six subjects, while the second contrasts humanities and mathematics subjects.
Communalities are, for each variable, the part of its variance that is explained by the common factors.
To estimate the communalities we need to sum the square of the factor loadings for each subject:
> lambda<-round(lambda,2)
> communality<-apply(lambda^2,1,sum)
> communality
[1] 0.4036 0.3848 0.3060 0.5218 0.5065 0.3688
Or, equivalently, communality<-diag(lambda%*%t(lambda))
These shows, for example, that the 40% of variance in Gaelic scores is explained by the two common factors. Of course, the larger the communality the better does the variable serve as an indicator of the associated factors.
To evaluate the goodness of fit of this model we can compute the residual correlation matrix ( ):
> R-lambda%*%t(lambda)
Gaelic English History Arithmetic Algebra Geometry
Gaelic 0.5964 0.0492 0.0668 -0.0042 0.0268 -0.0208
English 0.0492 0.6152 0.0272 0.0208 -0.0158 0.0332
History 0.0668 0.0272 0.6940 -0.0242 -0.0050 0.0012
Arithmetic -0.0042 0.0208 -0.0242 0.4782 0.0763 0.0328
Algebra 0.0268 -0.0158 -0.0050 0.0763 0.4935 0.0282
Geometry -0.0208 0.0332 0.0012 0.0328 0.0282 0.6312
Since the elements out of the diagonal are fairly small and close to zero we can conclude that the model fits adequately the data.
Athletics data
File AthleticsData.sav contains measurements over 9 different athletics disciplines on 1000 students:
3
1.
PINBALL
2.
BILLIARD
3.
GOLF
4.
1500 m
5.
2 Km row
6.
12 min RUN
7.
BENCH
8.
CURL
9.
MAX PUSHUP
The aim here is to reduce the dimension of the problem by measuring some latent factors that impact their performances.
The dataset has a SPSS format (extension .sav). To read the file we need to load an R-package that contains a function that allows this conversion.
> library(Hmisc)
> library(foreign)
> AthleticsData <- spss.get("AthleticsData.sav")
> x<-AthleticsData
>
> x[1:5,]
PINBALL BILLIARD GOLF X.1500M X.2KROW X.12MINTR BENCH
1 -1.1225055 0.009316132 -1.5267935 -0.9483176 -0.1647701 -0.05203922 1.3593056
2 0.3286001 -0.745125995 -0.8488870 0.6849068 0.1455623 0.13481553 -0.4906018
3 0.5442109 0.823572688 0.5519436 -0.6842024 -0.5152493 -0.24014598 -0.8188845
4 1.7282347 -0.142108710 0.9537609 0.9312700 -1.0275236 -0.89791136 -0.9732271
5 0.8650813 0.363277424 -0.5669886 0.9757308 1.2200180 0.24952087 1.2293106
CURL MAXPUSHU
1 -0.83766332 -0.04783271
2 0.22148232 0.38120977
3 -0.62012251 -1.13213981
4 -1.12976703 -0.33035037
5 -0.03462911 0.40703588
The R-function factanal performs maximum-likelihood factor analysis on a covariance (correlation) matrix or data matrix. It takes the following main arguments:
x: A formula or a numeric matrix or an object that can be
coerced to a numeric matrix.
factors: The number of factors to be fitted.
data: An optional data frame (or similar: see ‘model.frame’), used
only if ‘x’ is a formula. By default the variables are taken
from ‘environment(formula)’.
covmat: A covariance matrix, or a covariance list as returned by
‘cov.wt’. Of course, correlation matrices are covariance
matrices.
n.obs: The number of observations, used if ‘covmat’ is a covariance
matrix.
start: ‘NULL’ or a matrix of starting values, each column giving an
initial set of uniquenesses.
4
scores: Type of scores to produce, if any. The default is none,
‘"regression"’ gives Thompson's scores, ‘"Bartlett"’ given
Bartlett's weighted least-squares scores. Partial matching
allows these names to be abbreviated. rotation: character. ‘"none"’ or the name of a function to be used to
rotate the factors: it will be called with first argument the
loadings matrix, and should return a list with component
‘loadings’ giving the rotated loadings, or just the rotated
loadings.
To begin with, let’s analyze the AthleticsData with a 2 factor model.
> fit.2 <- factanal(x,factors=2,rotation="none")
> fit.2
Call: factanal(x = x, factors = 2, rotation = "none")
Uniquenesses:
PINBALL BILLIARD GOLF X.1500M X.2KROW X.12MINTR BENCH CURL
0.938 0.962 0.955 0.361 0.534 0.536 0.301 0.540
MAXPUSHU
0.560
Loadings:
Factor1 Factor2
PINBALL 0.249
BILLIARD 0.192
GOLF 0.206
X.1500M 0.793
X.2KROW 0.413 0.544
X.12MINTR 0.681
BENCH 0.813 -0.193
CURL 0.673
MAXPUSHU 0.545 0.379
Factor1 Factor2
SS loadings 1.734 1.579
Proportion Var 0.193 0.175
Cumulative Var 0.193 0.368
Test of the hypothesis that 2 factors are sufficient.
The chi square statistic is 652.4 on 19 degrees of freedom.
The p-value is 4.3e-126
Near the bottom of the output, we can see that the significance level of the χ2 fit statistic is very small. This indicates that the hypothesis that a 2 factor model fits the data is rejected. Since we are in a purely exploratory vein, let’s fit a 3 factor model.
> fit.3 <- factanal(x,factors=3,rotation="none")
> fit.3
Call: factanal(x = x, factors = 3, rotation = "none")
5
Uniquenesses:
PINBALL BILLIARD GOLF X.1500M X.2KROW X.12MINTR BENCH CURL
0.635 0.414 0.455 0.361 0.520 0.538 0.302 0.536
MAXPUSHU
0.540
Loadings:
Factor1 Factor2 Factor3
PINBALL 0.425 0.429
BILLIARD 0.443 0.624
GOLF 0.447 0.585
X.1500M 0.799
X.2KROW 0.408 0.496 -0.260
X.12MINTR 0.672
BENCH 0.729 -0.280 -0.297
CURL 0.605 -0.158 -0.270
MAXPUSHU 0.512 0.317 -0.312
Factor1 Factor2 Factor3
SS loadings 1.912 1.545 1.243
Proportion Var 0.212 0.172 0.138
Cumulative Var 0.212 0.384 0.522
Test of the hypothesis that 3 factors are sufficient.
The chi square statistic is 12.94 on 12 degrees of freedom.
The p-value is 0.373
These results are much more promising. Although the sample size is reasonably large, N = 1000, the significance level of .373 indicates that the hypothesis that a 3 factor model fits the data cannot be rejected. Changing from two factors to three has produced a huge improvement.
The output reports the uniquenesses i.e. the variances of the unique factors. As the algorithm fits the model using the correlation matrix, the communalities can be obtained as 1 minus the corresponding uniquenesses. For instance the communality for the variable pinball is 1-0.635=0.365.
The last table in the output reports the sum of squared loadings for each factor i.e.
0.425^2+0.443^2+…+0.512^2=1.912. It represents the part of the total variance that is explained by the first factor. If we divide it by the total variance (i.e. 9 in this case) we obtain the proportion of the total variance explained by the first factor. The first factor explains 21.2% of the total variance.
The unrotated factors do not have a clear interpretation. Some procedures have been developed to search automatically for a suitable rotation. For example, VARIMAX procedure attempts to find an orthogonal rotation that is close to simple structure by finding factors with few large loadings and as many near-zero loadings as possible. In order to improve the understanding of the problem let's try to rotate the axes with the VARIMAX procedure:
> fit.3 <- factanal(x,factors=3,rotation="varimax")
> fit.3
Call: factanal(x = x, factors = 3, rotation = "varimax")
Uniquenesses:
PINBALL BILLIARD GOLF X.1500M X.2KROW X.12MINTR BENCH CURL
0.635 0.414 0.455 0.361 0.520 0.538 0.302 0.536
MAXPUSHU
6
0.540
Loadings:
Factor1 Factor2 Factor3
PINBALL 0.131 0.590
BILLIARD 0.765
GOLF 0.735
X.1500M 0.779 -0.179
X.2KROW 0.585 0.372
X.12MINTR 0.678
BENCH -0.119 0.816 0.137
CURL 0.674
MAXPUSHU 0.433 0.522
Factor1 Factor2 Factor3
SS loadings 1.613 1.584 1.502
Proportion Var 0.179 0.176 0.167
Cumulative Var 0.179 0.355 0.522
Test of the hypothesis that 3 factors are sufficient.
The chi square statistic is 12.94 on 12 degrees of freedom.
The p-value is 0.373
As expected from the invariance of the factor model to orthogonal rotations the estimates of the communalities do not change after rotation.
We can "clean up" the factor pattern in several ways. One way is to hide small loadings, to reduce the visual clutter in the factor pattern. Another is to reduce the number of decimal places from 3 to 2. A third way is to sort the loadings to make the simple structure more obvious. The following command does all three:
> print(fit.3, digits = 2, cutoff = .2, sort = TRUE)
Call: factanal(x = x, factors = 3, rotation = "varimax")
Uniquenesses:
PINBALL BILLIARD GOLF X.1500M X.2KROW X.12MINTR BENCH CURL
0.64 0.41 0.46 0.36 0.52 0.54 0.30 0.54
MAXPUSHU
0.54
Loadings:
Factor1 Factor2 Factor3
X.1500M 0.78
X.2KROW 0.58 0.37
X.12MINTR 0.68
BENCH 0.82
CURL 0.67
MAXPUSHU 0.43 0.52
PINBALL 0.59
BILLIARD 0.76
GOLF 0.73
Factor1 Factor2 Factor3
SS loadings 1.61 1.58 1.50
Proportion Var 0.18 0.18 0.17
Cumulative Var 0.18 0.36 0.52
Test of the hypothesis that 3 factors are sufficient.
The chi square statistic is 12.94 on 12 degrees of freedom.
7
The p-value is 0.373
Now it is obvious that there are 3 factors. The traditional approach to naming factors is to:
Try do decide what construct is common to these variables
Examine the variables that load heavily on the factor
Name the factor after that construct
It seems that there are three factors. The first factor is something that is common to strong performance in a 1500 meter run, a 2000 meter row, and a 12 minute run. It looks like a good name for this factor is
"Endurance". The other two factors might be named "Strength", and "Hand-Eye Coordination".
Sometimes , we may want to calculate an individual's score on the latent variable(s). In factor analysis it is not straightforward, because the factors are random variables which have a probability distribution. There are various methods for obtaining predicted factor scores; the function factanal produces scores only if a data matrix is supplied and used. The first method is the regression method of Thomson, the second the weighted least squares method of Bartlett. scores_thomson<-factanal(x, factors = 3, scores = "regression")$scores scores_bartlett<-factanal(x, factors = 3, scores = "Bartlett")$scores
Example
File intel_test.txt shows correlations between scores of 75 children in 10 intelligence tests WPPSI:
X
1
: information
X
2
: vocabulary
X
3
: arithmetic
X
4
: similarities
X
5
: comprehension
X
6
: animal houses
X
7
: figures completion
X
8
: labyrinths
X
9
: geometric design
X
10
: block design
> cor.m<-as.matrix(read.table("c:\\temp\\intel_test.txt"))
>
> cor.m
V1 V2 V3 V4 V5 V6 V7 V8 V9 V10
1 1.000 0.755 0.592 0.532 0.627 0.460 0.407 0.387 0.461 0.459
2 0.755 1.000 0.644 0.520 0.617 0.497 0.511 0.417 0.406 0.583
3 0.592 0.644 1.000 0.388 0.529 0.449 0.436 0.428 0.412 0.602
4 0.532 0.528 0.388 1.000 0.475 0.442 0.280 0.214 0.361 0.424
5 0.627 0.617 0.529 0.475 1.000 0.398 0.373 0.372 0.350 0.433
6 0.460 0.497 0.449 0.442 0.398 1.000 0.545 0.446 0.366 0.575
7 0.407 0.511 0.436 0.280 0.373 0.545 1.000 0.542 0.308 0.590
8 0.387 0.417 0.428 0.214 0.372 0.446 0.542 1.000 0.375 0.654
9 0.461 0.406 0.412 0.361 0.355 0.366 0.308 0.375 1.000 0.502
10 0.459 0.583 0.602 0.424 0.433 0.575 0.590 0.654 0.502 1.000
By looking at the correlation matrix one can see a strong correlation between the 10 tests: all the correlation values are positive and mostly varies between 0.4-0.6.
8
Factor analysis according to a maximum likelihood approach:
> res<-factanal(covmat=cor.m,factors=2,n.obs=75,rotation="none")
>
> res
Call: factanal(factors = 2, covmat = cor.m, n.obs = 75, rotation = "none")
Uniquenesses:
V1 V2 V3 V4 V5 V6 V7 V8 V9 V10
0.215 0.249 0.452 0.622 0.482 0.553 0.534 0.481 0.679 0.177
Loadings:
Factor1 Factor2
[1,] 0.789 -0.403
[2,] 0.834 -0.234
[3,] 0.740
[4,] 0.587 -0.185
[5,] 0.676 -0.247
[6,] 0.654 0.140
[7,] 0.641 0.235
[8,] 0.630 0.351
[9,] 0.564
[10,] 0.807 0.414
Factor1 Factor2
SS loadings 4.872 0.685
Proportion Var 0.487 0.069
Cumulative Var 0.487 0.556
Test of the hypothesis that 2 factors are sufficient.
The chi square statistic is 16.51 on 26 degrees of freedom.
The p-value is 0.923
Record the percentage of variability in each variable that is explained by the model (communalities):
> round(apply(res$loadings^2,1,sum),3)
[1] 0.785 0.751 0.548 0.378 0.518 0.447 0.466 0.519 0.321 0.823
Rotate the factors with VARIMAX. Such a rotation works on the factor loadings increasing the differences between lower weights, letting them converge to zero, and the higher weights, letting them converge to one.
> res.rot<-factanal(covmat=cor.m,factors=2,n.obs=75,rotation="varimax")
>
> res.rot
Call: factanal(factors = 2, covmat = cor.m, n.obs = 75, rotation = "varimax")
Uniquenesses:
V1 V2 V3 V4 V5 V6 V7 V8 V9 V10
0.215 0.249 0.452 0.622 0.482 0.553 0.534 0.481 0.679 0.177
9
Loadings:
Factor1 Factor2
[1,] 0.852 0.245
[2,] 0.769 0.399
[3,] 0.563 0.481
[4,] 0.555 0.266
[5,] 0.662 0.281
[6,] 0.382 0.549
[7,] 0.308 0.609
[8,] 0.220 0.686
[9,] 0.375 0.424
[10,] 0.307 0.854
Factor1 Factor2
SS loadings 2.904 2.653
Proportion Var 0.290 0.265
Cumulative Var 0.290 0.556
Test of the hypothesis that 2 factors are sufficient.
The chi square statistic is 16.51 on 26 degrees of freedom.
The p-value is 0.923
>
10