International Journal of Data Mining and Bioinformatics (in press)
advertisement

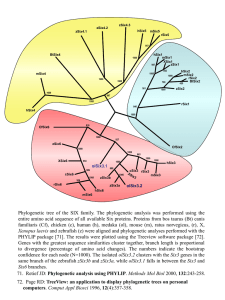
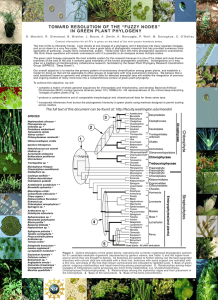
International Journal of Data Mining and Bioinformatics (in press) Molecular Phylogeny Analysis Using Correlation Distance And Spectral Distance. Anu Sabarish R, Tessamma Thomas Abstract: A wide range of methods with or without sequence alignment have been used to study molecular phylogeny for information on the evolution of species. Two approaches to construct the phylogenetic tree using, i) direct correlation of protein sequences and ii) difference between the Discrete Fourier Transform coefficients, are described. The proposed methods use a transformation where each amino acid is represented by its electron-ion interaction potential (EIIP) value. Phylogenetic tree of two mammalian orders, primates and cetacea, are generated based on FitchMargoliash, Neighbor Joining and UPGMA methods and compared. The phylogenetic tree of evolutionary relationships thus obtained can be used for comparison of species and gene sequences. The information thus gathered provide meaningful insights into the pattern and process of evolution which will help researchers in developing new breeds of animals and plants. Keywords: Genomic Signal Processing, Discrete Fourier Transform, Discrete Wavelet Transform, Correlation, Electron Ion Interaction Potential, Phylogenetic tree. Acceptance Date: 14 Aug 2012