nph12765-sup-0001-FigsS1-S3-TableS1
advertisement

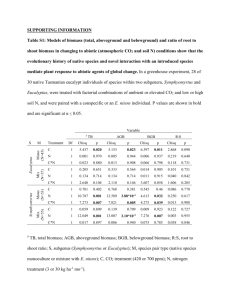
Supporting Information Methods S1, Table S1, Figs S1–S3 Species richness of arbuscular mycorrhizal fungi: associations with grassland plant richness and biomass Inga Hiiesalu, Meelis Pärtel, John Davison, Pille Gerhold, Madis Metsis, Mari Moora, Maarja Öpik, Martti Vasar, Martin Zobel and Scott D. Wilson Methods S1 The number of reads varied considerably between samples in the AMF and belowground plant data sets (see start of Results section). Variable sequence counts could indicate that different proportions of the true diversity were recorded in different samples. This in turn could influence relationships between diversity and explanatory variables if sequence counts varied systematically, or if they add noise to the relationships. An approach that is commonly used to overcome this is to rarefy or otherwise downsample data sets to a uniform or similar sampling depth (de Cárcer et al., 2011). However the merits of this approach depend on the reasons for between-sample variation. Rarefaction or other types of downsampling may remove infrequent taxa from a data set, and if there are natural as opposed to technical explanations for variations in sample depth, then this may itself bias diversity estimates. In this study, we inferred the richness of plants above- and belowground from fixed volumes (10x10x10 cm). Meanwhile, belowground plant and fungal richness was estimated from DNA samples extracted from a subsample of roots constituting a fixed mass of roots — identical for all samples (100 mg) — expected to yield representative estimates of the diversity of organisms present in the root samples. Thus, our approach was based on sampling a constant volume, and differences in the numbers of sequences were anticipated. Nonetheless, we used rarefaction to test whether variation in sampling depth within the data sets qualitatively influenced our results and thus whether our findings could have been influenced by technical artifacts. We used rarefaction to downsample the AMF and belowground plant data sets using the function rrarefy() from the R package vegan (Oksanen et al., 2013). We randomly resampled the median number of sequences per sample (264 sequences for AMF and 64 sequences for plants). Median sample numbers perform better than the minimum sample numbers for high-throughput sequencing datasets (de Cárcer et al., 2011). Following this, we used Z-tests to compare the correlation coefficients derived from analysis of the original and rarefied data sets. Rarefaction did not change any of the correlations significantly (P values between 0.41 and 1; Table S1), although the coefficients tended to be slightly lower. We believe this is mostly due to a loss of statistical power, since the rarefaction resulted in considerable loss of data (35% of sequences in the AMF dataset and and 70% in the belowground plant dataset). References de Cárcer DA de, Denman SE, McSweeney C, Morrison M. 2011. Evaluation of subsamplingbased normalization strategies for tagged high-throughput sequencing data sets from gut microbiomes. Applied and Environmental Microbiology 77: 8795–8798. Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2013. Vegan: Community ecology package. R package version 2.0-7. http://CRAN.R project.org/package=vegan Table S1 Summary of the Z-test which compares the r values of the original (unrarefied) and rarefied data sets Original Rarefied Difference of Correlation r1 z1 r2 z2 z1-z2 SE Z-score P of Z AMF richness vs AG plant richess 0.28 0.29 0.28 0.29 0 0.18 0 1 AMF richness vs BG plant richness 0.48 0.51 0.43 0.46 0.06 0.18 0.34 0.73 BG plant richness vs BG plant biomass 0.32 0.33 0.18 0.18 0.15 0.18 0.82 0.41 BG plant richness vs total plant biomass 0.31 0.32 0.17 0.17 0.15 0.18 0.81 0.42 AMF richness vs BG plant biomass -0.29 0.30 -0.20 0.20 0.1 0.18 0.52 0.60 AMF richness vs total plant biomass -0.31 0.32 -0.23 0.23 0.09 0.18 0.47 0.64 AMF, arbuscular mycorrhizal fungi; AG, aboveground; BG, belowground; SE, standard error. Fig. S1 Partial correlation between arbuscular mycorrhizal fungal (AMF) richness and (a) aboveground plant richness; and (b) belowground plant richness (the sum of species rooted in a sample and species detected from root samples using 454-sequencing of the trnL (UAA) gene). Sequence data were rarefied to median number of sequences prior to analysis. Residuals of the x and y variables are plotted in order to account for the effects of other measured variables and spatial autocorrelation. Fig. S2 Partial correlation between plant (a) aboveground richness and belowground biomass; (b) belowground richness and belowground biomass; and (c) belowground richness and total biomass. Belowground plant richness is the sum of species rooted in a sample and species detected from root samples using 454-sequencing of the trnL (UAA) gene. Sequence data were rarefied to median number of sequences prior to analysis. Total biomass is the sum of above- and belowground plant biomass. Residuals of the x and y variables are plotted in order to account for the effects of other measured variables and spatial autocorrelation. Fig. S3 Partial correlation between arbuscular mycorrhizal fungal (AMF) richness (number of SSU rRNA gene based virtual taxa) and (a) belowground plant biomass, and (b) total plant biomass (sum of above- and belowground plant biomass). Sequence data rarefied to median number of sequences prior to analysis. Residuals of the x and y variables are plotted in order to account for the effects of other measured variables and spatial autocorrelation.